Accurate quantification of transcriptome from RNA-Seq data by effective length normalization
- PMID: 21059678
- PMCID: PMC3025570
- DOI: 10.1093/nar/gkq1015
Accurate quantification of transcriptome from RNA-Seq data by effective length normalization
Abstract
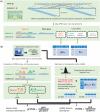
We propose a novel, efficient and intuitive approach of estimating mRNA abundances from the whole transcriptome shotgun sequencing (RNA-Seq) data. Our method, NEUMA (Normalization by Expected Uniquely Mappable Area), is based on effective length normalization using uniquely mappable areas of gene and mRNA isoform models. Using the known transcriptome sequence model such as RefSeq, NEUMA pre-computes the numbers of all possible gene-wise and isoform-wise informative reads: the former being sequences mapped to all mRNA isoforms of a single gene exclusively and the latter uniquely mapped to a single mRNA isoform. The results are used to estimate the effective length of genes and transcripts, taking experimental distributions of fragment size into consideration. Quantitative RT-PCR based on 27 randomly selected genes in two human cell lines and computer simulation experiments demonstrated superior accuracy of NEUMA over other recently developed methods. NEUMA covers a large proportion of genes and mRNA isoforms and offers a measure of consistency ('consistency coefficient') for each gene between an independently measured gene-wise level and the sum of the isoform levels. NEUMA is applicable to both paired-end and single-end RNA-Seq data. We propose that NEUMA could make a standard method in quantifying gene transcript levels from RNA-Seq data.
Figures






References
-
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. - PubMed
-
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. - PubMed

