Conflicts targeting epigenetic systems and their resolution by cell death: novel concepts for methyl-specific and other restriction systems
- PMID: 21059708
- PMCID: PMC2993543
- DOI: 10.1093/dnares/dsq027
Conflicts targeting epigenetic systems and their resolution by cell death: novel concepts for methyl-specific and other restriction systems
Abstract
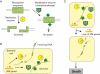
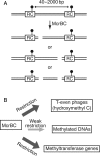
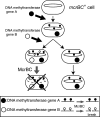
Epigenetic modification of genomic DNA by methylation is important for defining the epigenome and the transcriptome in eukaryotes as well as in prokaryotes. In prokaryotes, the DNA methyltransferase genes often vary, are mobile, and are paired with the gene for a restriction enzyme. Decrease in a certain epigenetic methylation may lead to chromosome cleavage by the partner restriction enzyme, leading to eventual cell death. Thus, the pairing of a DNA methyltransferase and a restriction enzyme forces an epigenetic state to be maintained within the genome. Although restriction enzymes were originally discovered for their ability to attack invading DNAs, it may be understood because such DNAs show deviation from this epigenetic status. DNAs with epigenetic methylation, by a methyltransferase linked or unlinked with a restriction enzyme, can also be the target of DNases, such as McrBC of Escherichia coli, which was discovered because of its methyl-specific restriction. McrBC responds to specific genome methylation systems by killing the host bacterial cell through chromosome cleavage. Evolutionary and genomic analysis of McrBC homologues revealed their mobility and wide distribution in prokaryotes similar to restriction-modification systems. These findings support the hypothesis that this family of methyl-specific DNases evolved as mobile elements competing with specific genome methylation systems through host killing. These restriction systems clearly demonstrate the presence of conflicts between epigenetic systems.
Figures



References
-
- Lyko F., Ramsahoye B.H., Kashevsky H., et al. Mammalian (cytosine-5) methyltransferases cause genomic DNA methylation and lethality in Drosophila. Nat. Genet. 1999;23:363–6. - PubMed
-
- Kimura H., Suetake I., Tajima S. Exogenous expression of mouse Dnmt3 induces apoptosis in Xenopus early embryos. J. Biochem. 2002;131:933–41. - PubMed
-
- Roos W.P., Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006;12:440–50. - PubMed
-
- Warren R.A. Modified bases in bacteriophage DNAs. Annu. Rev. Microbiol. 1980;34:137–58. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

