The centrosome neither persistently leads migration nor determines the site of axonogenesis in migrating neurons in vivo
- PMID: 21059852
- PMCID: PMC2983064
- DOI: 10.1083/jcb.201004154
The centrosome neither persistently leads migration nor determines the site of axonogenesis in migrating neurons in vivo
Erratum in
- J Cell Biol. 2010 Dec 27;191(7):1413
Abstract
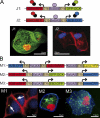
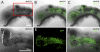
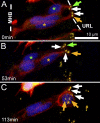
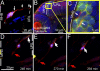
The position of the centrosome ahead of the nucleus has been considered crucial for coordinating neuronal migration in most developmental situations. The proximity of the centrosome has also been correlated with the site of axonogenesis in certain differentiating neurons. Despite these positive correlations, accumulating experimental findings appear to negate a universal role of the centrosome in determining where an axon forms, or in leading the migration of neurons. To further examine this controversy in an in vivo setting, we have generated cell type-specific multi-cistronic gene expression to monitor subcellular dynamics in the developing zebrafish cerebellum. We show that migration of rhombic lip-derived neurons is characterized by a centrosome that does not persistently lead the nucleus, but which is instead regularly overtaken by the nucleus. In addition, axonogenesis is initiated during the onset of neuronal migration and occurs independently of centrosome proximity. These in vivo data reveal a new temporal orchestration of organelle dynamics and provide important insights into the variation in intracellular processes during vertebrate brain differentiation.
Figures





References
-
- Asakawa K., Suster M.L., Mizusawa K., Nagayoshi S., Kotani T., Urasaki A., Kishimoto Y., Hibi M., Kawakami K. 2008. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl. Acad. Sci. USA. 105:1255–1260 10.1073/pnas.0704963105 - DOI - PMC - PubMed
-
- Bellion A., Baudoin J.P., Alvarez C., Bornens M., Métin C. 2005. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J. Neurosci. 25:5691–5699 10.1523/JNEUROSCI.1030-05.2005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

