Gain of MYC underlies recurrent trisomy of the MYC chromosome in acute promyelocytic leukemia
- PMID: 21059853
- PMCID: PMC2989761
- DOI: 10.1084/jem.20091071
Gain of MYC underlies recurrent trisomy of the MYC chromosome in acute promyelocytic leukemia
Abstract
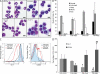
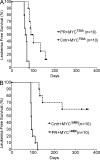
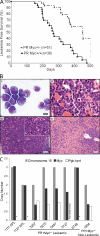
Gain of chromosome 8 is the most common chromosomal gain in human acute myeloid leukemia (AML). It has been hypothesized that gain of the MYC protooncogene is of central importance in trisomy 8, but the experimental data to support this are limited and controversial. In a mouse model of promyelocytic leukemia in which the MRP8 promoter drives expression of the PML-RARA fusion gene in myeloid cells, a Myc allele is gained in approximately two-thirds of cases as a result of trisomy for mouse chromosome 15. We used this model to test the idea that MYC underlies acquisition of trisomy in AML. We used a retroviral vector to drive expression of wild-type, hypermorphic, or hypomorphic MYC in bone marrow that expressed the PML-RARA transgene. MYC retroviruses cooperated in myeloid leukemogenesis and suppressed gain of chromosome 15. When the PML-RARA transgene was expressed in a Myc haploinsufficient background, we observed selection for increased copies of the wild-type Myc allele concomitant with leukemic transformation. In addition, we found that human myeloid leukemias with trisomy 8 have increased MYC. These data show that gain of MYC can contribute to the pathogenic effect of the most common trisomy of human AML.
Figures



Similar articles
-
Co-operative leukemogenesis in acute myeloid leukemia and acute promyelocytic leukemia reveals C/EBPα as a common target of TRIB1 and PML/RARA.Haematologica. 2016 Oct;101(10):1228-1236. doi: 10.3324/haematol.2015.138503. Epub 2016 Jul 6. Haematologica. 2016. PMID: 27390356 Free PMC article.
-
Recurring chromosomal abnormalities in leukemia in PML-RARA transgenic mice parallel human acute promyelocytic leukemia.Blood. 2002 Apr 15;99(8):2985-91. doi: 10.1182/blood.v99.8.2985. Blood. 2002. PMID: 11929790
-
Typical, atypical and cryptic t(15;17)(q24;q21) (PML::RARA) observed in acute promyelocytic leukemia: A retrospective review of 831 patients with concurrent chromosome and PML::RARA dual-color dual-fusion FISH studies.Genes Chromosomes Cancer. 2022 Oct;61(10):629-634. doi: 10.1002/gcc.23070. Epub 2022 Jun 10. Genes Chromosomes Cancer. 2022. PMID: 35639830
-
Isochromosome der(17)(q10)t(15;17) in acute promyelocytic leukemia resulting in an additional copy of the RARA-PML fusion gene: report of 4 cases and review of the literature.Acta Haematol. 2010;123(3):162-70. doi: 10.1159/000294959. Epub 2010 Mar 11. Acta Haematol. 2010. PMID: 20224268 Review.
-
Function of PML-RARA in Acute Promyelocytic Leukemia.Adv Exp Med Biol. 2024;1459:321-339. doi: 10.1007/978-3-031-62731-6_14. Adv Exp Med Biol. 2024. PMID: 39017850 Review.
Cited by
-
PML-RARα co-operates with Sox4 in acute myeloid leukemia development in mice.Haematologica. 2013 Mar;98(3):424-7. doi: 10.3324/haematol.2011.057067. Epub 2012 Nov 9. Haematologica. 2013. PMID: 23144197 Free PMC article.
-
New insights into the troubles of aneuploidy.Annu Rev Cell Dev Biol. 2012;28:189-214. doi: 10.1146/annurev-cellbio-101011-155807. Epub 2012 Jul 9. Annu Rev Cell Dev Biol. 2012. PMID: 22804579 Free PMC article. Review.
-
Overexpression of the long non-coding RNA PVT1 is correlated with leukemic cell proliferation in acute promyelocytic leukemia.J Hematol Oncol. 2015 Nov 6;8:126. doi: 10.1186/s13045-015-0223-4. J Hematol Oncol. 2015. PMID: 26545364 Free PMC article.
-
Tumor suppressor function of Gata2 in acute promyelocytic leukemia.Blood. 2021 Sep 30;138(13):1148-1161. doi: 10.1182/blood.2021011758. Blood. 2021. PMID: 34125173 Free PMC article.
-
Trisomy 8 Associated Clonal Cytopenia Featured With Acquired Auto-Inflammation and Its Response to JAK Inhibitors.Front Med (Lausanne). 2022 Apr 25;9:895965. doi: 10.3389/fmed.2022.895965. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35547205 Free PMC article.
References
-
- Akagi T., Shih L.Y., Kato M., Kawamata N., Yamamoto G., Sanada M., Okamoto R., Miller C.W., Liang D.C., Ogawa S., Koeffler H.P. 2009. Hidden abnormalities and novel classification of t(15;17) acute promyelocytic leukemia (APL) based on genomic alterations. Blood. 113:1741–1748 10.1182/blood-2007-12-130260 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

