Force-induced formation and propagation of adhesion nanodomains in living fungal cells
- PMID: 21059927
- PMCID: PMC2996417
- DOI: 10.1073/pnas.1013893107
Force-induced formation and propagation of adhesion nanodomains in living fungal cells
Abstract
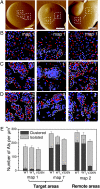
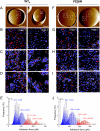
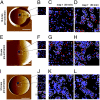
Understanding how cell adhesion proteins form adhesion domains is a key challenge in cell biology. Here, we use single-molecule atomic force microscopy (AFM) to demonstrate the force-induced formation and propagation of adhesion nanodomains in living fungal cells, focusing on the covalently anchored cell-wall protein Als5p from Candida albicans. We show that pulling on single adhesins with AFM tips terminated with specific antibodies triggers the formation of adhesion domains of 100-500 nm and that the force-induced nanodomains propagate over the entire cell surface. Control experiments (with cells lacking Als5p, single-site mutation in the protein, bare tips, and tips modified with irrelevant antibodies) demonstrate that Als5p nanodomains result from protein redistribution triggered by force-induced conformational changes in the initially probed proteins, rather than from nonspecific cell-wall perturbations. Als5p remodeling is independent of cellular metabolic activity because heat-killed cells show the same behavior as live cells. Using AFM and fluorescence microscopy, we also find that nanodomains are formed within ∼30 min and migrate at a speed of ∼20 nm·min(-1), indicating that domain formation and propagation are slow, time-dependent processes. These results demonstrate that mechanical stimuli can trigger adhesion nanodomains in fungal cells and suggest that the force-induced clustering of adhesins may be a mechanism for activating cell adhesion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat Rev Neurosci. 2007;8:11–20. - PubMed
-
- Weber C, Fraemohs L, Dejana E. The role of junctional adhesion molecules in vascular inflammation. Nat Rev Immunol. 2007;7:467–477. - PubMed
-
- Gray-Owen SD, Blumberg RS. CEACAM1: Contact-dependent control of immunity. Nat Rev Immunol. 2006;6:433–446. - PubMed
-
- Verstrepen KJ, Reynolds TB, Fink GR. Origins of variation in the fungal cell surface. Nat Rev Microbiol. 2004;2:533–540. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

