Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import
- PMID: 21059946
- PMCID: PMC2996643
- DOI: 10.1073/pnas.1010095107
Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import
Abstract
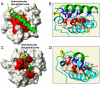
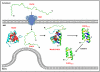
Several proteins of the mitochondrial intermembrane space are targeted by internal targeting signals. A class of such proteins with α-helical hairpin structure bridged by two intramolecular disulfides is trapped by a Mia40-dependent oxidative process. Here, we describe the oxidative folding mechanism underpinning this process by an exhaustive structural characterization of the protein in all stages and as a complex with Mia40. Two consecutive induced folding steps are at the basis of the protein-trapping process. In the first one, Mia40 functions as a molecular chaperone assisting α-helical folding of the internal targeting signal of the substrate. Subsequently, in a Mia40-independent manner, folding of the second substrate helix is induced by the folded targeting signal functioning as a folding scaffold. The Mia40-induced folding pathway provides a proof of principle for the general concept that internal targeting signals may operate as a folding nucleus upon compartment-specific activation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. - PubMed
-
- Lu H, et al. Functional TIM10 chaperone assembly is redox-regulated in vivo. J Biol Chem. 2004;279:18952–18958. - PubMed
-
- Webb CT, et al. Crystal structure of the mitochondrial chaperone TIM9.10 reveals a six-bladed alpha-propeller. Mol Cell. 2006;21:123–133. - PubMed
-
- Banci L, et al. A structural-dynamical characterization of human Cox17. J Biol Chem. 2008;283:7912–7920. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

