Multidrug-resistant enterococci lack CRISPR-cas
- PMID: 21060735
- PMCID: PMC2975353
- DOI: 10.1128/mBio.00227-10
Multidrug-resistant enterococci lack CRISPR-cas
Abstract
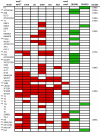
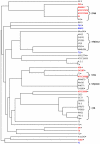
Clustered, regularly interspaced short palindromic repeats (CRISPR) provide bacteria and archaea with sequence-specific, acquired defense against plasmids and phage. Because mobile elements constitute up to 25% of the genome of multidrug-resistant (MDR) enterococci, it was of interest to examine the codistribution of CRISPR and acquired antibiotic resistance in enterococcal lineages. A database was built from 16 Enterococcus faecalis draft genome sequences to identify commonalities and polymorphisms in the location and content of CRISPR loci. With this data set, we were able to detect identities between CRISPR spacers and sequences from mobile elements, including pheromone-responsive plasmids and phage, suggesting that CRISPR regulates the flux of these elements through the E. faecalis species. Based on conserved locations of CRISPR and CRISPR-cas loci and the discovery of a new CRISPR locus with associated functional genes, CRISPR3-cas, we screened additional E. faecalis strains for CRISPR content, including isolates predating the use of antibiotics. We found a highly significant inverse correlation between the presence of a CRISPR-cas locus and acquired antibiotic resistance in E. faecalis, and examination of an additional eight E. faecium genomes yielded similar results for that species. A mechanism for CRISPR-cas loss in E. faecalis was identified. The inverse relationship between CRISPR-cas and antibiotic resistance suggests that antibiotic use inadvertently selects for enterococcal strains with compromised genome defense.
Figures

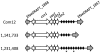

References
-
- Top J., Willems R., Bonten M. 2008. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol. Med. Microbiol. 52:297–308 - PubMed
-
- Werner G., Coque T. M., Hammerum A. M., Hope R., Hryniewicz W., Johnson A., Klare I., Kristinsson K. G., Leclercq R., Lester C. H., Lillie M., Novais C., Olsson-Liljequist B., Peixe L. V., Sadowy E., Simonsen G. S., Top J., Vuopio-Varkila J., Willems R. J., Witte W., Woodford N. 2008. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro. Surveill. 13(47):p ii=19046 - PubMed
-
- Hidron A. I., Edwards J. R., Patel J., Horan T. C., Sievert D. M., Pollock D. A., Fridkin S. K. 2008. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect. Control Hosp. Epidemiol. 29:996–1011 - PubMed
-
- Kak V., Chow J. W. 2002. Acquired antibiotic resistances in enterococci, p. 355–383 In Gilmore M. S., The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC
-
- Weigel L. M., Clewell D. B., Gill S. R., Clark N. C., McDougal L. K., Flannagan S. E., Kolonay J. F., Shetty J., Killgore G. E., Tenover F. C. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569–1571 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

