Retention and loss of RNA interference pathways in trypanosomatid protozoans
- PMID: 21060810
- PMCID: PMC2965760
- DOI: 10.1371/journal.ppat.1001161
Retention and loss of RNA interference pathways in trypanosomatid protozoans
Abstract
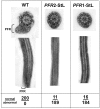
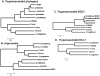
RNA interference (RNAi) pathways are widespread in metaozoans but the genes required show variable occurrence or activity in eukaryotic microbes, including many pathogens. While some Leishmania lack RNAi activity and Argonaute or Dicer genes, we show that Leishmania braziliensis and other species within the Leishmania subgenus Viannia elaborate active RNAi machinery. Strong attenuation of expression from a variety of reporter and endogenous genes was seen. As expected, RNAi knockdowns of the sole Argonaute gene implicated this protein in RNAi. The potential for functional genetics was established by testing RNAi knockdown lines lacking the paraflagellar rod, a key component of the parasite flagellum. This sets the stage for the systematic manipulation of gene expression through RNAi in these predominantly diploid asexual organisms, and may also allow selective RNAi-based chemotherapy. Functional evolutionary surveys of RNAi genes established that RNAi activity was lost after the separation of the Leishmania subgenus Viannia from the remaining Leishmania species, a divergence associated with profound changes in the parasite infectious cycle and virulence. The genus Leishmania therefore offers an accessible system for testing hypothesis about forces that may select for the loss of RNAi during evolution, such as invasion by viruses, changes in genome plasticity mediated by transposable elements and gene amplification (including those mediating drug resistance), and/or alterations in parasite virulence.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



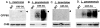

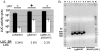
Similar articles
-
An RNA Interference (RNAi) Toolkit and Its Utility for Functional Genetic Analysis of Leishmania (Viannia).Genes (Basel). 2022 Dec 28;14(1):93. doi: 10.3390/genes14010093. Genes (Basel). 2022. PMID: 36672832 Free PMC article.
-
The Evolutionary Loss of RNAi Key Determinants in Kinetoplastids as a Multiple Sporadic Phenomenon.J Mol Evol. 2017 Mar;84(2-3):104-115. doi: 10.1007/s00239-017-9780-1. Epub 2017 Feb 16. J Mol Evol. 2017. PMID: 28210761 Free PMC article.
-
The structure and repertoire of small interfering RNAs in Leishmania (Viannia) braziliensis reveal diversification in the trypanosomatid RNAi pathway.Mol Microbiol. 2013 Feb;87(3):580-93. doi: 10.1111/mmi.12117. Epub 2012 Dec 26. Mol Microbiol. 2013. PMID: 23217017 Free PMC article.
-
Protozomics: trypanosomatid parasite genetics comes of age.Nat Rev Genet. 2003 Jan;4(1):11-9. doi: 10.1038/nrg980. Nat Rev Genet. 2003. PMID: 12509749 Review.
-
The evolution of RNAi as a defence against viruses and transposable elements.Philos Trans R Soc Lond B Biol Sci. 2009 Jan 12;364(1513):99-115. doi: 10.1098/rstb.2008.0168. Philos Trans R Soc Lond B Biol Sci. 2009. PMID: 18926973 Free PMC article. Review.
Cited by
-
CRISPR/Cas9-Induced Disruption of Paraflagellar Rod Protein 1 and 2 Genes in Trypanosoma cruzi Reveals Their Role in Flagellar Attachment.mBio. 2015 Jul 21;6(4):e01012. doi: 10.1128/mBio.01012-15. mBio. 2015. PMID: 26199333 Free PMC article.
-
Novel features of a PIWI-like protein homolog in the parasitic protozoan Leishmania.PLoS One. 2012;7(12):e52612. doi: 10.1371/journal.pone.0052612. Epub 2012 Dec 21. PLoS One. 2012. PMID: 23285111 Free PMC article.
-
Metagenomic sequencing suggests a diversity of RNA interference-like responses to viruses across multicellular eukaryotes.PLoS Genet. 2018 Jul 30;14(7):e1007533. doi: 10.1371/journal.pgen.1007533. eCollection 2018 Jul. PLoS Genet. 2018. PMID: 30059538 Free PMC article.
-
Glucose Transporters and Virulence in Leishmania mexicana.mSphere. 2018 Aug 1;3(4):e00349-18. doi: 10.1128/mSphere.00349-18. mSphere. 2018. PMID: 30068561 Free PMC article.
-
Concentration of 2'C-methyladenosine triphosphate by Leishmania guyanensis enables specific inhibition of Leishmania RNA virus 1 via its RNA polymerase.J Biol Chem. 2018 Apr 27;293(17):6460-6469. doi: 10.1074/jbc.RA117.001515. Epub 2018 Mar 6. J Biol Chem. 2018. PMID: 29511088 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

