Fidelity variants of RNA dependent RNA polymerases uncover an indirect, mutagenic activity of amiloride compounds
- PMID: 21060812
- PMCID: PMC2965762
- DOI: 10.1371/journal.ppat.1001163
Fidelity variants of RNA dependent RNA polymerases uncover an indirect, mutagenic activity of amiloride compounds
Abstract
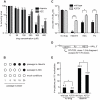
In a screen for RNA mutagen resistance, we isolated a high fidelity RNA dependent RNA polymerase (RdRp) variant of Coxsackie virus B3 (CVB3). Curiously, this variant A372V is also resistant to amiloride. We hypothesize that amiloride has a previously undescribed mutagenic activity. Indeed, amiloride compounds increase the mutation frequencies of CVB3 and poliovirus and high fidelity variants of both viruses are more resistant to this effect. We hypothesize that this mutagenic activity is mediated through alterations in intracellular ions such as Mg²+ and Mn²+, which in turn increase virus mutation frequency by affecting RdRp fidelity. Furthermore, we show that another amiloride-resistant RdRp variant, S299T, is completely resistant to this mutagenic activity and unaffected by changes in ion concentrations. We show that RdRp variants resist the mutagenic activity of amiloride via two different mechanisms: 1) increased fidelity that generates virus populations presenting lower basal mutation frequencies or 2) resisting changes in divalent cation concentrations that affect polymerase fidelity. Our results uncover a new antiviral approach based on mutagenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Viral Polymerase-Helicase Complexes Regulate Replication Fidelity To Overcome Intracellular Nucleotide Depletion.J Virol. 2015 Nov;89(22):11233-44. doi: 10.1128/JVI.01553-15. Epub 2015 Aug 26. J Virol. 2015. PMID: 26311883 Free PMC article.
-
Ribavirin-resistant variants of foot-and-mouth disease virus: the effect of restricted quasispecies diversity on viral virulence.J Virol. 2014 Apr;88(8):4008-20. doi: 10.1128/JVI.03594-13. Epub 2014 Jan 22. J Virol. 2014. PMID: 24453363 Free PMC article.
-
Understanding the Mechanism of the Broad-Spectrum Antiviral Activity of Favipiravir (T-705): Key Role of the F1 Motif of the Viral Polymerase.J Virol. 2017 May 26;91(12):e00487-17. doi: 10.1128/JVI.00487-17. Print 2017 Jun 15. J Virol. 2017. PMID: 28381577 Free PMC article.
-
Ribavirin and lethal mutagenesis of poliovirus: molecular mechanisms, resistance and biological implications.Virus Res. 2005 Feb;107(2):173-81. doi: 10.1016/j.virusres.2004.11.007. Virus Res. 2005. PMID: 15649563 Review.
-
Nidovirus RNA polymerases: Complex enzymes handling exceptional RNA genomes.Virus Res. 2017 Apr 15;234:58-73. doi: 10.1016/j.virusres.2017.01.023. Epub 2017 Feb 6. Virus Res. 2017. PMID: 28174054 Free PMC article. Review.
Cited by
-
Porcine Reproductive and Respiratory Syndrome Modified Live Virus Vaccine: A "Leaky" Vaccine with Debatable Efficacy and Safety.Vaccines (Basel). 2021 Apr 9;9(4):362. doi: 10.3390/vaccines9040362. Vaccines (Basel). 2021. PMID: 33918580 Free PMC article. Review.
-
Mutational robustness of an RNA virus influences sensitivity to lethal mutagenesis.J Virol. 2012 Mar;86(5):2869-73. doi: 10.1128/JVI.05712-11. Epub 2011 Dec 21. J Virol. 2012. PMID: 22190724 Free PMC article.
-
Inter and intra-host diversity of RSV in hematopoietic stem cell transplant adults with normal and delayed viral clearance.Virus Evol. 2023 Dec 28;10(1):vead086. doi: 10.1093/ve/vead086. eCollection 2024. Virus Evol. 2023. PMID: 38361816 Free PMC article.
-
Changes in protein domains outside the catalytic site of the bacteriophage Qβ replicase reduce the mutagenic effect of 5-azacytidine.J Virol. 2014 Sep;88(18):10480-7. doi: 10.1128/JVI.00979-14. Epub 2014 Jun 25. J Virol. 2014. PMID: 24965463 Free PMC article.
-
Deep sequencing analysis of viral infection and evolution allows rapid and detailed characterization of viral mutant spectrum.Bioinformatics. 2015 Jul 1;31(13):2141-50. doi: 10.1093/bioinformatics/btv101. Epub 2015 Feb 19. Bioinformatics. 2015. PMID: 25701575 Free PMC article.
References
-
- Horisberger JD. Amiloride-sensitive Na channels. Curr Opin Cell Biol. 1998;10:443–449. - PubMed
-
- Premkumar A, Horan CR, Gage PW. Dengue virus M protein C-terminal peptide (DVM-C) forms ion channels. J Membr Biol. 2005;204:33–38. - PubMed
-
- Ewart GD, Mills K, Cox GB, Gage PW. Amiloride derivatives block ion channel activity and enhancement of virus-like particle budding caused by HIV-1 protein Vpu. Eur Biophys J. 2002;31:26–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

