Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite
- PMID: 21060817
- PMCID: PMC2965767
- DOI: 10.1371/journal.ppat.1001165
Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite
Abstract
The molecular mechanisms underlying transcriptional regulation in apicomplexan parasites remain poorly understood. Recently, the Apicomplexan AP2 (ApiAP2) family of DNA binding proteins was identified as a major class of transcriptional regulators that are found across all Apicomplexa. To gain insight into the regulatory role of these proteins in the malaria parasite, we have comprehensively surveyed the DNA-binding specificities of all 27 members of the ApiAP2 protein family from Plasmodium falciparum revealing unique binding preferences for the majority of these DNA binding proteins. In addition to high affinity primary motif interactions, we also observe interactions with secondary motifs. The ability of a number of ApiAP2 proteins to bind multiple, distinct motifs significantly increases the potential complexity of the transcriptional regulatory networks governed by the ApiAP2 family. Using these newly identified sequence motifs, we infer the trans-factors associated with previously reported plasmodial cis-elements and provide evidence that ApiAP2 proteins modulate key regulatory decisions at all stages of parasite development. Our results offer a detailed view of ApiAP2 DNA binding specificity and take the first step toward inferring comprehensive gene regulatory networks for P. falciparum.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

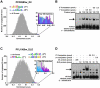
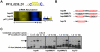


References
-
- WHO. World Malaria Report 2009 2009.
-
- Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. - PubMed
-
- Hall N, Karras M, Raine JD, Carlton JM, Kooij TW, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. - PubMed
-
- Lasonder E, Ishihama Y, Andersen JS, Vermunt AM, Pain A, et al. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002;419:537–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

