A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer
- PMID: 21060830
- PMCID: PMC2966402
- DOI: 10.1371/journal.pone.0013735
A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer
Abstract
Background: To date, there are no highly sensitive and specific minimally invasive biomarkers for detection of breast cancer at an early stage. The occurrence of circulating microRNAs (miRNAs) in blood components (including serum and plasma) has been repeatedly observed in cancer patients as well as healthy controls. Because of the significance of miRNA in carcinogenesis, circulating miRNAs in blood may be unique biomarkers for early and minimally invasive diagnosis of human cancers. The objective of this pilot study was to discover a panel of circulating miRNAs as potential novel breast cancer biomarkers.
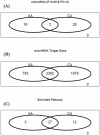
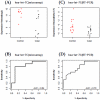
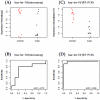
Methodology/principal findings: Using microarray-based expression profiling followed by Real-Time quantitative Polymerase Cycle Reaction (RT-qPCR) validation, we compared the levels of circulating miRNAs in plasma samples from 20 women with early stage breast cancer (10 Caucasian American (CA) and 10 African American (AA)) and 20 matched healthy controls (10 CAs and 10 AAs). Using the significance level of p<0.05 constrained by at least two-fold expression change as selection criteria, we found that 31 miRNAs were differentially expressed in CA study subjects (17 up and 14 down) and 18 miRNAs were differentially expressed in AA study subjects (9 up and 9 down). Interestingly, only 2 differentially expressed miRNAs overlapped between CA and AA study subjects. Using receiver operational curve (ROC) analysis, we show that not only up-regulated but also down-regulated miRNAs can discriminate patients with breast cancer from healthy controls with reasonable sensitivity and specificity. To further explore the potential roles of these circulating miRNAs in breast carcinogenesis, we applied pathway-based bioinformatics exploratory analysis and predicted a number of significantly enriched pathways which are predicted to be regulated by these circulating miRNAs, most of which are involved in critical cell functions, cancer development and progression.
Conclusions: Our observations from this pilot study suggest that the altered levels of circulating miRNAs might have great potential to serve as novel, noninvasive biomarkers for early detection of breast cancer.
Conflict of interest statement
Figures




References
-
- Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, et al. Herceptin Adjuvant (HERA) Trial Study Team. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–1672. - PubMed
-
- Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, et al. American Society of Clinical Oncology. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

