An assay for RNA oxidation induced abasic sites using the Aldehyde Reactive Probe
- PMID: 21062214
- PMCID: PMC3058411
- DOI: 10.3109/10715762.2010.535529
An assay for RNA oxidation induced abasic sites using the Aldehyde Reactive Probe
Erratum in
- Free Radic Res. 2011 May;45(5):642. Song, Han [corrected to Han, Song]
Abstract
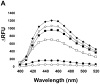
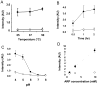
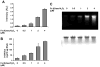
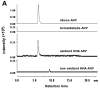
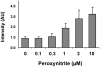
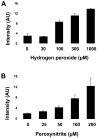
There have been several reports describing elevation of oxidized RNA in ageing or age-related diseases, however RNA oxidation has been assessed solely based on 8-hydroxy-guanosine levels. In this study, Aldehyde Reactive Probe (ARP), which was originally developed to detect DNA abasic sites, was used to assess RNA oxidation. It was found that ARP reacted with depurinated tRNA(Phe) or chemically synthesized RNA containing abasic sites quantitatively to as little as 10 fmoles, indicating that abasic RNA is recognized by ARP. RNA oxidized by Fenton-type reactions, γ-irradiation or peroxynitrite increased ARP reactivity dose-dependently, indicating that ARP is capable of monitoring oxidized RNA mediated by reactive oxygen species or reactive nitrogen species. Furthermore, oxidative stress increased levels of ARP reactive RNA in cultured cells. These results indicate the versatility of the assay method for biologically relevant oxidation of RNA. Thus, this study developed a sensitive assay for analysis of oxidized RNA.
Figures



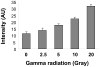

Similar articles
-
Quantification of oxidized levels of specific RNA species using an aldehyde reactive probe.Anal Biochem. 2011 Oct 1;417(1):142-8. doi: 10.1016/j.ab.2011.05.038. Epub 2011 May 30. Anal Biochem. 2011. PMID: 21693097 Free PMC article.
-
Synthesis and damage specificity of a novel probe for the detection of abasic sites in DNA.Biochemistry. 1993 Aug 17;32(32):8276-83. doi: 10.1021/bi00083a031. Biochemistry. 1993. PMID: 8347625
-
Estimation of the Level of Abasic Sites in Plant mRNA Using Aldehyde Reactive Probe.Methods Mol Biol. 2022;2526:125-134. doi: 10.1007/978-1-0716-2469-2_9. Methods Mol Biol. 2022. PMID: 35657516
-
Role of oxidant species in aging.Curr Med Chem. 2004 May;11(9):1105-12. doi: 10.2174/0929867043365341. Curr Med Chem. 2004. PMID: 15134509 Review.
-
Peroxynitrite induced signaling pathways in plant response to non-proteinogenic amino acids.Planta. 2020 Jun 13;252(1):5. doi: 10.1007/s00425-020-03411-4. Planta. 2020. PMID: 32535658 Free PMC article. Review.
Cited by
-
Cadmium Stress Leads to Rapid Increase in RNA Oxidative Modifications in Soybean Seedlings.Front Plant Sci. 2018 Jan 9;8:2219. doi: 10.3389/fpls.2017.02219. eCollection 2017. Front Plant Sci. 2018. PMID: 29375597 Free PMC article.
-
Quantification of oxidized levels of specific RNA species using an aldehyde reactive probe.Anal Biochem. 2011 Oct 1;417(1):142-8. doi: 10.1016/j.ab.2011.05.038. Epub 2011 May 30. Anal Biochem. 2011. PMID: 21693097 Free PMC article.
-
METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation.Mol Cell. 2019 Jun 20;74(6):1278-1290.e9. doi: 10.1016/j.molcel.2019.03.040. Epub 2019 Apr 25. Mol Cell. 2019. PMID: 31031083 Free PMC article.
-
The Impact of Oxidative Stress on Ribosomes: From Injury to Regulation.Cells. 2019 Nov 2;8(11):1379. doi: 10.3390/cells8111379. Cells. 2019. PMID: 31684095 Free PMC article. Review.
-
Markers of oxidant stress that are clinically relevant in aging and age-related disease.Mech Ageing Dev. 2013 Mar;134(3-4):139-57. doi: 10.1016/j.mad.2013.02.008. Epub 2013 Feb 18. Mech Ageing Dev. 2013. PMID: 23428415 Free PMC article. Review.
References
-
- Liu J, Head E, Gharib AM, Yuan W, Ingersoll RT, Hagen TM, Cotman CW, Ames BN. Memory loss in old rats is associated with brain mitochondrial decay and RNA/DNA oxidation: partial reversal by feeding acetyl-L-carnitine and/or R-alpha -lipoic acid. Proc Natl Acad Sci U S A. 2002;99:2356–2361. - PMC - PubMed
-
- Martinet W, de Meyer GR, Herman AG, Kockx MM. Reactive oxygen species induce RNA damage in human atherosclerosis. European J Clin Investigat. 2004;34:323–327. - PubMed
-
- Nunomura A, Chiba S, Kosaka K, Takeda A, Castellani RJ, Smith MA, Perry G. Neuronal RNA oxidation is a prominent feature of dementia with Lewy bodies.[erratum appears in Neuroreport. 2003 Feb 10;14(2):293] Neuroreport. 2002;13:2035–2039. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
