A genome-wide association scan on estrogen receptor-negative breast cancer
- PMID: 21062454
- PMCID: PMC3046434
- DOI: 10.1186/bcr2772
A genome-wide association scan on estrogen receptor-negative breast cancer
Abstract
Introduction: Breast cancer is a heterogeneous disease and may be characterized on the basis of whether estrogen receptors (ER) are expressed in the tumour cells. ER status of breast cancer is important clinically, and is used both as a prognostic indicator and treatment predictor. In this study, we focused on identifying genetic markers associated with ER-negative breast cancer risk.
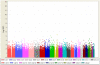
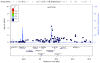
Methods: We conducted a genome-wide association analysis of 285,984 single nucleotide polymorphisms (SNPs) genotyped in 617 ER-negative breast cancer cases and 4,583 controls. We also conducted a genome-wide pathway analysis on the discovery dataset using permutation-based tests on pre-defined pathways. The extent of shared polygenic variation between ER-negative and ER-positive breast cancers was assessed by relating risk scores, derived using ER-positive breast cancer samples, to disease state in independent, ER-negative breast cancer cases.
Results: Association with ER-negative breast cancer was not validated for any of the five most strongly associated SNPs followed up in independent studies (1,011 ER-negative breast cancer cases, 7,604 controls). However, an excess of small P-values for SNPs with known regulatory functions in cancer-related pathways was found (global P = 0.052). We found no evidence to suggest that ER-negative breast cancer shares a polygenic basis to disease with ER-positive breast cancer.
Conclusions: ER-negative breast cancer is a distinct breast cancer subtype that merits independent analyses. Given the clinical importance of this phenotype and the likelihood that genetic effect sizes are small, greater sample sizes and further studies are required to understand the etiology of ER-negative breast cancers.
Figures








References
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. - PMC - PubMed
-
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. - PubMed
-
- Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, Hankinson SE, Hutchinson A, Wang Z, Yu K, Chatterjee N, Garcia-Closas M, Gonzalez-Bosquet J, Prokunina-Olsson L, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Diver R, Prentice R, Jackson R, Kooperberg C, Chlebowski R, Lissowska J. et al.A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–584. - PMC - PubMed
-
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R, Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le Marchand L, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, Fletcher O. et al.Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

