Mannose trimming is required for delivery of a glycoprotein from EDEM1 to XTP3-B and to late endoplasmic reticulum-associated degradation steps
- PMID: 21062743
- PMCID: PMC3020737
- DOI: 10.1074/jbc.M110.154849
Mannose trimming is required for delivery of a glycoprotein from EDEM1 to XTP3-B and to late endoplasmic reticulum-associated degradation steps
Abstract
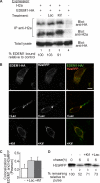
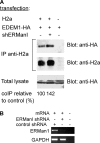
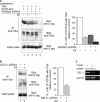
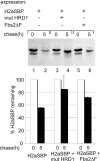
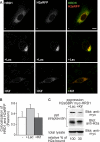
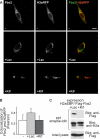
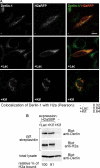
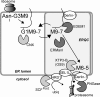
Although the trimming of α1,2-mannose residues from precursor N-linked oligosaccharides is an essential step in the delivery of misfolded glycoproteins to endoplasmic reticulum (ER)-associated degradation (ERAD), the exact role of this trimming is unclear. EDEM1 was initially suggested to bind N-glycans after mannose trimming, a role presently ascribed to the lectins OS9 and XTP3-B, because of their in vitro affinities for trimmed oligosaccharides. We have shown before that ER mannosidase I (ERManI) is required for the trimming and concentrates together with the ERAD substrate and ERAD machinery in the pericentriolar ER-derived quality control compartment (ERQC). Inhibition of mannose trimming prevents substrate accumulation in the ERQC. Here, we show that the mannosidase inhibitor kifunensine or ERManI knockdown do not affect binding of an ERAD substrate glycoprotein to EDEM1. In contrast, substrate association with XTP3-B and with the E3 ubiquitin ligases HRD1 and SCF(Fbs2) was inhibited. Consistently, whereas the ERAD substrate partially colocalized upon proteasomal inhibition with EDEM1, HRD1, and Fbs2 at the ERQC, colocalization was repressed by mannosidase inhibition in the case of the E3 ligases but not for EDEM1. Interestingly, association and colocalization of the substrate with Derlin-1 was independent of mannose trimming. The HRD1 adaptor protein SEL1L had been suggested to play a role in N-glycan-dependent substrate delivery to OS9 and XTP3-B. However, substrate association with XTP3-B was still dependent on mannose trimming upon SEL1L knockdown. Our results suggest that mannose trimming enables delivery of a substrate glycoprotein from EDEM1 to late ERAD steps through association with XTP3-B.
Figures

Similar articles
-
Bypass of glycan-dependent glycoprotein delivery to ERAD by up-regulated EDEM1.Mol Biol Cell. 2011 Nov;22(21):3945-54. doi: 10.1091/mbc.E10-12-0944. Epub 2011 Sep 14. Mol Biol Cell. 2011. PMID: 21917589 Free PMC article.
-
Mannosidase IA is in Quality Control Vesicles and Participates in Glycoprotein Targeting to ERAD.J Mol Biol. 2016 Aug 14;428(16):3194-3205. doi: 10.1016/j.jmb.2016.04.020. Epub 2016 Apr 21. J Mol Biol. 2016. PMID: 27108681
-
Endoplasmic reticulum lectin XTP3-B inhibits endoplasmic reticulum-associated degradation of a misfolded α1-antitrypsin variant.FEBS J. 2013 Mar;280(6):1563-75. doi: 10.1111/febs.12157. Epub 2013 Feb 28. FEBS J. 2013. PMID: 23356641
-
Quality control of glycoprotein folding and ERAD: the role of N-glycan handling, EDEM1 and OS-9.Histochem Cell Biol. 2017 Feb;147(2):269-284. doi: 10.1007/s00418-016-1513-9. Epub 2016 Nov 1. Histochem Cell Biol. 2017. PMID: 27803995 Review.
-
Glycoprotein folding, quality control and ER-associated degradation.Curr Opin Struct Biol. 2009 Oct;19(5):515-23. doi: 10.1016/j.sbi.2009.06.004. Epub 2009 Jul 17. Curr Opin Struct Biol. 2009. PMID: 19616933 Review.
Cited by
-
Glycoprotein folding and quality-control mechanisms in protein-folding diseases.Dis Model Mech. 2014 Mar;7(3):331-41. doi: 10.1242/dmm.014589. Dis Model Mech. 2014. PMID: 24609034 Free PMC article. Review.
-
Abnormal expression of ER quality control and ER associated degradation proteins in the dorsolateral prefrontal cortex in schizophrenia.Schizophr Res. 2018 Jul;197:484-491. doi: 10.1016/j.schres.2018.02.010. Epub 2018 Feb 26. Schizophr Res. 2018. PMID: 29496332 Free PMC article.
-
Arabidopsis Class I α-Mannosidases MNS4 and MNS5 Are Involved in Endoplasmic Reticulum-Associated Degradation of Misfolded Glycoproteins.Plant Cell. 2014 Apr;26(4):1712-1728. doi: 10.1105/tpc.114.123216. Epub 2014 Apr 15. Plant Cell. 2014. PMID: 24737672 Free PMC article.
-
A shared endoplasmic reticulum-associated degradation pathway involving the EDEM1 protein for glycosylated and nonglycosylated proteins.J Biol Chem. 2013 Jan 25;288(4):2167-78. doi: 10.1074/jbc.M112.438275. Epub 2012 Dec 11. J Biol Chem. 2013. PMID: 23233672 Free PMC article.
-
The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology.Physiol Rev. 2012 Apr;92(2):537-76. doi: 10.1152/physrev.00027.2011. Physiol Rev. 2012. PMID: 22535891 Free PMC article. Review.
References
-
- Aebi M., Bernasconi R., Clerc S., Molinari M. (2010) Trends Biochem. Sci. 35, 74–82 - PubMed
-
- Hebert D. N., Bernasconi R., Molinari M. (2010) Semin. Cell Dev. Biol. 21, 526–532 - PubMed
-
- Lederkremer G. Z. (2009) Curr. Opin. Struct. Biol. 19, 515–523 - PubMed
-
- Määttänen P., Gehring K., Bergeron J. J., Thomas D. Y. (2010) Semin. Cell Dev. Biol. 21, 500–511 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

