Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha} (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease
- PMID: 21062749
- PMCID: PMC3020752
- DOI: 10.1074/jbc.M110.145870
Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha} (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease
Abstract
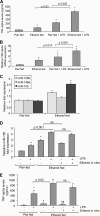
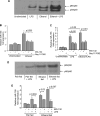
Activation of Kupffer cells (KCs) by gut-derived lipopolysaccharide (LPS) and Toll-Like Receptors 4 (TLR4)-LPS-mediated increase in TNFα production has a central role in the pathogenesis of alcoholic liver disease. Micro-RNA (miR)-125b, miR-146a, and miR-155 can regulate inflammatory responses to LPS. Here we evaluated the involvement of miRs in alcohol-induced macrophage activation. Chronic alcohol treatment in vitro resulted in a time-dependent increase in miR-155 but not miR-125b or miR-146a levels in RAW 264.7 macrophages. Furthermore, alcohol pretreatment augmented LPS-induced miR-155 expression in macrophages. We found a linear correlation between alcohol-induced increase in miR-155 and TNFα induction. In a mouse model of alcoholic liver disease, we found a significant increase in both miR-155 levels and TNFα production in isolated KCs when compared with pair-fed controls. The mechanistic role of miR-155 in TNFα regulation was indicated by decreased TNFα levels in alcohol-treated macrophages after inhibition of miR-155 and by increased TNFα production after miR-155 overexpression, respectively. We found that miR-155 affected TNFα mRNA stability because miR-155 inhibition decreased whereas miR-155 overexpression increased TNFα mRNA half-life. Using the NF-κB inhibitors, MG-132 or Bay11-7082, we demonstrated that NF-κB activation mediated the up-regulation of miR-155 by alcohol in KCs. In conclusion, our novel data demonstrate that chronic alcohol consumption increases miR-155 in macrophages via NF-κB and the increased miR-155 contributes to alcohol-induced elevation in TNFα production via increased mRNA stability.
Figures





References
-
- Baltimore D., Boldin M. P., O'Connell R. M., Rao D. S., Taganov K. D. (2008) Nat. Immunol. 9, 839–845 - PubMed
-
- Bi Y., Liu G., Yang R. (2009) J. Cell. Physiol. 218, 467–472 - PubMed
-
- Tili E., Michaille J. J., Cimino A., Costinean S., Dumitru C. D., Adair B., Fabbri M., Alder H., Liu C. G., Calin G. A., Croce C. M. (2007) J. Immunol. 179, 5082–5089 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

