Phospho.ELM: a database of phosphorylation sites--update 2011
- PMID: 21062810
- PMCID: PMC3013696
- DOI: 10.1093/nar/gkq1104
Phospho.ELM: a database of phosphorylation sites--update 2011
Abstract
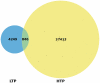
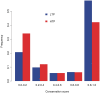
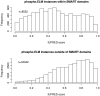
The Phospho.ELM resource (http://phospho.elm.eu.org) is a relational database designed to store in vivo and in vitro phosphorylation data extracted from the scientific literature and phosphoproteomic analyses. The resource has been actively developed for more than 7 years and currently comprises 42,574 serine, threonine and tyrosine non-redundant phosphorylation sites. Several new features have been implemented, such as structural disorder/order and accessibility information and a conservation score. Additionally, the conservation of the phosphosites can now be visualized directly on the multiple sequence alignment used for the score calculation. Finally, special emphasis has been put on linking to external resources such as interaction networks and other databases.
Figures

References
-
- Cantin GT, Yi W, Lu B, Park SK, Xu T, Lee JD, Yates JR. Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J. Proteome Res. 2008;7:1346–1351. - PubMed
-
- Mann M, Ong SE, Gronborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. - PubMed
-
- Ballif BA, Villen J, Beausoleil SA, Schwartz D, Gygi SP. Phosphoproteomic analysis of the developing mouse brain. Mol Cell Proteomics. 2004;3:1093–1101. - PubMed
-
- Lemeer S, Heck AJ. The phosphoproteomics data explosion. Curr. Opin. Chem. Biol. 2009;13:414–420. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

