Analysis of thioester-containing proteins during the innate immune response of Drosophila melanogaster
- PMID: 21063077
- PMCID: PMC3031515
- DOI: 10.1159/000321554
Analysis of thioester-containing proteins during the innate immune response of Drosophila melanogaster
Abstract
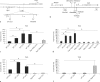
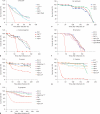
Thioester-containing proteins (TEPs) are conserved proteins among insects that are thought to be involved in innate immunity. In Drosophila, the Tep family is composed of 6 genes named Tep1-Tep6. In this study, we investigated the phylogeny, expression pattern and roles of these genes in the host defense of Drosophila. Protostomian Tep genes are clustered in 3 distinct branches, 1 of which is specific to mosquitoes. Most D. melanogaster Tep genes are expressed in hemocytes, can be induced in the fat body, and are expressed in specific regions of the hypodermis. This expression pattern is consistent with a role in innate immunity. However, we find that TEP1, TEP2, and TEP4 are not strictly required in the body cavity to fight several bacterial and fungal infections. One possibility is that Drosophila TEPs act redundantly or that their absence can be compensated by other components of the immune response. TEPs may thus provide a subtle selective advantage during evolution. Alternatively, they may be required in host defense against specific as yet unidentified natural pathogens of Drosophila.
Copyright © 2010 S. Karger AG, Basel.
Figures





Similar articles
-
The distinct function of Tep2 and Tep6 in the immune defense of Drosophila melanogaster against the pathogen Photorhabdus.Virulence. 2017 Nov 17;8(8):1668-1682. doi: 10.1080/21505594.2017.1330240. Epub 2017 Jun 2. Virulence. 2017. PMID: 28498729 Free PMC article.
-
Thioester-Containing Proteins 2 and 4 Affect the Metabolic Activity and Inflammation Response in Drosophila.Infect Immun. 2018 Apr 23;86(5):e00810-17. doi: 10.1128/IAI.00810-17. Print 2018 May. Infect Immun. 2018. PMID: 29463615 Free PMC article.
-
The Drosophila Thioester containing Protein-4 participates in the induction of the cellular immune response to the pathogen Photorhabdus.Dev Comp Immunol. 2017 Nov;76:200-208. doi: 10.1016/j.dci.2017.06.008. Epub 2017 Jun 19. Dev Comp Immunol. 2017. PMID: 28642050
-
Pattern recognition receptors in Drosophila immune responses.Dev Comp Immunol. 2020 Jan;102:103468. doi: 10.1016/j.dci.2019.103468. Epub 2019 Aug 17. Dev Comp Immunol. 2020. PMID: 31430488 Review.
-
Functional role of thioester-containing proteins in the Drosophila anti-pathogen immune response.Dev Comp Immunol. 2023 Feb;139:104578. doi: 10.1016/j.dci.2022.104578. Epub 2022 Oct 18. Dev Comp Immunol. 2023. PMID: 36270515 Review.
Cited by
-
The Role of Mosquito Hemocytes in Viral Infections.Viruses. 2022 Sep 20;14(10):2088. doi: 10.3390/v14102088. Viruses. 2022. PMID: 36298644 Free PMC article. Review.
-
New levels of transcriptome complexity at upper thermal limits in wild Drosophila revealed by exon expression analysis.Genetics. 2013 Nov;195(3):809-30. doi: 10.1534/genetics.113.156224. Epub 2013 Sep 3. Genetics. 2013. PMID: 24002645 Free PMC article.
-
Arthropod Innate Immune Systems and Vector-Borne Diseases.Biochemistry. 2017 Feb 21;56(7):907-918. doi: 10.1021/acs.biochem.6b00870. Epub 2017 Feb 8. Biochemistry. 2017. PMID: 28072517 Free PMC article. Review.
-
RNAseq Analysis of the Drosophila Response to the Entomopathogenic Nematode Steinernema.G3 (Bethesda). 2017 Jun 7;7(6):1955-1967. doi: 10.1534/g3.117.041004. G3 (Bethesda). 2017. PMID: 28450373 Free PMC article.
-
Transgenic Expression of the Anti-parasitic Factor TEP1 in the Malaria Mosquito Anopheles gambiae.PLoS Pathog. 2017 Jan 17;13(1):e1006113. doi: 10.1371/journal.ppat.1006113. eCollection 2017 Jan. PLoS Pathog. 2017. PMID: 28095489 Free PMC article.
References
-
- Sansonetti PJ. The innate signaling of dangers and the dangers of innate signaling. Nat Immunol. 2006;7:1237–1242. - PubMed
-
- Sottrup-Jensen L, Hansen HF, Mortensen SB, Petersen TE, Magnusson S. Sequence location of the reactive thiol ester in human alpha 2-macroglobulin. FEBS Lett. 1981;123:145–148. - PubMed
-
- Medzhitov R, Janeway CA., Jr Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296:298–300. - PubMed
-
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

