Tamoxifen-independent recombination in the RIP-CreER mouse
- PMID: 21063464
- PMCID: PMC2965077
- DOI: 10.1371/journal.pone.0013533
Tamoxifen-independent recombination in the RIP-CreER mouse
Abstract
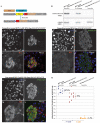
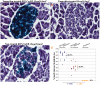
Background: The inducible Cre-lox system is a valuable tool to study gene function in a spatial and time restricted fashion in mouse models. This strategy relies on the limited background activity of the modified Cre recombinase (CreER) in the absence of its inducer, the competitive estrogen receptor ligand, tamoxifen. The RIP-CreER mouse (Tg (Ins2-cre/Esr1) 1Dam) is among the few available β-cell specific CreER mouse lines and thus it has been often used to manipulate gene expression in the insulin-producing cells of the endocrine pancreas.
Principal findings: Here, we report the detection of tamoxifen-independent Cre activity as early as 2 months of age in RIP-CreER mice crossed with three distinct reporter strains.
Significance: Evidence of Cre-mediated recombination of floxed alleles even in the absence of tamoxifen administration should warrant cautious use of this mouse for the study of pancreatic β-cells.
Conflict of interest statement
Figures


Similar articles
-
A mouse model with tamoxifen-inducible thyrocyte-specific cre recombinase activity.Genesis. 2014 Apr;52(4):333-40. doi: 10.1002/dvg.22740. Epub 2014 Jan 15. Genesis. 2014. PMID: 24395757
-
Tamoxifen-independent recombination of reporter genes limits lineage tracing and mosaic analysis using CreERT2 lines.Transgenic Res. 2020 Feb;29(1):53-68. doi: 10.1007/s11248-019-00177-8. Epub 2019 Oct 22. Transgenic Res. 2020. PMID: 31641921 Free PMC article.
-
Highly B lymphocyte-specific tamoxifen inducible transgene expression of CreER T2 by using the LC-1 locus BAC vector.Genesis. 2009 Nov;47(11):729-35. doi: 10.1002/dvg.20549. Genesis. 2009. PMID: 19621440
-
A review of tamoxifen administration regimen optimization for Cre/loxp system in mouse bone study.Biomed Pharmacother. 2023 Sep;165:115045. doi: 10.1016/j.biopha.2023.115045. Epub 2023 Jun 28. Biomed Pharmacother. 2023. PMID: 37379643 Review.
-
Endothelial-Specific Cre Mouse Models.Arterioscler Thromb Vasc Biol. 2018 Nov;38(11):2550-2561. doi: 10.1161/ATVBAHA.118.309669. Arterioscler Thromb Vasc Biol. 2018. PMID: 30354251 Free PMC article. Review.
Cited by
-
An Inducible Diabetes Mellitus Murine Model Based on MafB Conditional Knockout under MafA-Deficient Condition.Int J Mol Sci. 2020 Aug 5;21(16):5606. doi: 10.3390/ijms21165606. Int J Mol Sci. 2020. PMID: 32764399 Free PMC article.
-
Conditional gene expression systems in the transgenic rat brain.BMC Biol. 2012 Sep 3;10:77. doi: 10.1186/1741-7007-10-77. BMC Biol. 2012. PMID: 22943311 Free PMC article.
-
Benefits and Caveats in the Use of Retinal Pigment Epithelium-Specific Cre Mice.Int J Mol Sci. 2024 Jan 20;25(2):1293. doi: 10.3390/ijms25021293. Int J Mol Sci. 2024. PMID: 38279294 Free PMC article. Review.
-
iRAGu: A Novel Inducible and Reversible Mouse Model for Ubiquitous Recombinase Activity.Front Immunol. 2017 Nov 10;8:1525. doi: 10.3389/fimmu.2017.01525. eCollection 2017. Front Immunol. 2017. PMID: 29176980 Free PMC article.
-
Insulin-positive ductal cells do not migrate into preexisting islets during pregnancy.Exp Mol Med. 2021 Apr;53(4):605-614. doi: 10.1038/s12276-021-00593-z. Epub 2021 Apr 5. Exp Mol Med. 2021. PMID: 33820959 Free PMC article.
References
-
- Feil R. Conditional somatic mutagenesis in the mouse using site-specific recombinases. Handb Exp Pharmacol. 2007. pp. 3–28. - PubMed
-
- Joyner AL, Zervas M. Genetic inducible fate mapping in mouse: establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev Dyn. 2006;235:2376–2385. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

