Synthetic biology: putting synthesis into biology
- PMID: 21064036
- PMCID: PMC3057768
- DOI: 10.1002/wsbm.104
Synthetic biology: putting synthesis into biology
Abstract
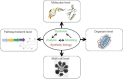
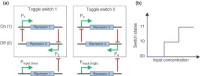
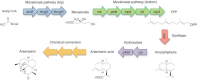
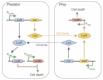
The ability to manipulate living organisms is at the heart of a range of emerging technologies that serve to address important and current problems in environment, energy, and health. However, with all its complexity and interconnectivity, biology has for many years been recalcitrant to engineering manipulations. The recent advances in synthesis, analysis, and modeling methods have finally provided the tools necessary to manipulate living systems in meaningful ways and have led to the coining of a field named synthetic biology. The scope of synthetic biology is as complicated as life itself--encompassing many branches of science and across many scales of application. New DNA synthesis and assembly techniques have made routine customization of very large DNA molecules. This in turn has allowed the incorporation of multiple genes and pathways. By coupling these with techniques that allow for the modeling and design of protein functions, scientists have now gained the tools to create completely novel biological machineries. Even the ultimate biological machinery--a self-replicating organism--is being pursued at this moment. The aim of this article is to dissect and organize these various components of synthetic biology into a coherent picture.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

