An aldol-based build/couple/pair strategy for the synthesis of medium- and large-sized rings: discovery of macrocyclic histone deacetylase inhibitors
- PMID: 21067169
- PMCID: PMC3004530
- DOI: 10.1021/ja105119r
An aldol-based build/couple/pair strategy for the synthesis of medium- and large-sized rings: discovery of macrocyclic histone deacetylase inhibitors
Abstract
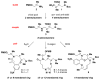
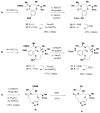
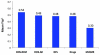
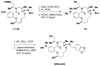
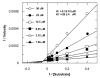
An aldol-based build/couple/pair (B/C/P) strategy was applied to generate a collection of stereochemically and skeletally diverse small molecules. In the build phase, a series of asymmetric syn- and anti-aldol reactions were performed to produce four stereoisomers of a Boc-protected γ-amino acid. In addition, both stereoisomers of O-PMB-protected alaninol were generated to provide a chiral amine coupling partner. In the couple step, eight stereoisomeric amides were synthesized by coupling the chiral acid and amine building blocks. The amides were subsequently reduced to generate the corresponding secondary amines. In the pair phase, three different reactions were employed to enable intramolecular ring-forming processes: nucleophilic aromatic substitution (S(N)Ar), Huisgen [3+2] cycloaddition, and ring-closing metathesis (RCM). Despite some stereochemical dependencies, the ring-forming reactions were optimized to proceed with good to excellent yields, providing a variety of skeletons ranging in size from 8- to 14-membered rings. Scaffolds resulting from the RCM pairing reaction were diversified on the solid phase to yield a 14 400-membered library of macrolactams. Screening of this library led to the discovery of a novel class of histone deacetylase inhibitors, which display mixed enzyme inhibition, and led to increased levels of acetylation in a primary mouse neuron culture. The development of stereo-structure/activity relationships was made possible by screening all 16 stereoisomers of the macrolactams produced through the aldol-based B/C/P strategy.
Figures

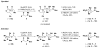



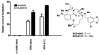

References
-
- Lovering F, Bikker J, Humblet C. J. Med. Chem. 2009;52:6752–6756. - PubMed
-
- Ganesan A. Curr. Opin. Chem. Biol. 2008;12:306–317. - PubMed
-
- Hübel K, Leβmann T, Waldmann H. Chem. Soc. Rev. 2008;37:1361–1374. - PubMed
- Koch MA, Schuffenhauer A, Scheck M, Wetzel S, Casaulta M, Odermatt A, Ertl P, Waldmann H. Proc. Natl. Acad. Sci. 2005;102:17272–17277. - PMC - PubMed
- Bauer RA, Wurst JM, Tan DS. Curr. Opin. Chem. Biol. 2010;14:308–314. - PMC - PubMed
- Marcaurelle LA, Johannes CW. Prog. Drug Res. 2008;66(187):189–216. - PubMed
- Goess BC, Hannoush RN, Chan LK, Kirchhausen T, Shair M. J. Am. Chem. Soc. 2006;128:5391–5403. - PMC - PubMed
- Pelish HE, Westwood NJ, Feng Y, Kirchhausen T, Shair MD. J. Am. Chem. Soc. 2001;123:6740–6741. - PubMed
- Marcaurelle LA, Johannes C, Yohannes D, Tillotson BP, Mann D. Bioorg. Med. Chem. Lett. 2009;19:2500–2503. - PubMed
-
- Zhang QS, Lu HJ, Curran DP. J. Am. Chem. Soc. 2004;126:36–37. - PubMed
- Curran DP, Zhang Q, Richard C, Lu H, Gudipathi V, Wilcox CS. J. Am. Chem. Soc. 2006;128:9561–9573. - PubMed
- Dandapani S, Jeske M, Curran DP. J. Org. Chem. 2005;70:9447–9462. - PubMed
- Wrona IE, Lowe JT, Turbyville TJ, Johnson TR, Beignet J, Beutler JA, Panek JS. J. Org. Chem. 2008;74:1897–1916. - PMC - PubMed
-
- Neilson TE, Schreiber SL. Angew. Chem. Int. Ed. 2007;74:48–56.
- Uchida T, Rodriguez M, Schreiber SL. Org. Lett. 2009;11:1559–1562. - PMC - PubMed
- Schreiber SL. Nature. 2009;457:153–154. - PubMed
- Luo T, Schreiber SL. J. Am. Chem. Soc. 2009;131:5667–5674. - PMC - PubMed
- Pizzirani D, Kaya T, Clemons PA, Schreiber SL. Org. Lett. 2010;12:2822–2825. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

