A first genome assembly of the barley fungal pathogen Pyrenophora teres f. teres
- PMID: 21067574
- PMCID: PMC3156948
- DOI: 10.1186/gb-2010-11-11-r109
A first genome assembly of the barley fungal pathogen Pyrenophora teres f. teres
Abstract
Background: Pyrenophora teres f. teres is a necrotrophic fungal pathogen and the cause of one of barley's most important diseases, net form of net blotch. Here we report the first genome assembly for this species based solely on short Solexa sequencing reads of isolate 0-1. The assembly was validated by comparison to BAC sequences, ESTs, orthologous genes and by PCR, and complemented by cytogenetic karyotyping and the first genome-wide genetic map for P. teres f. teres.
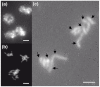
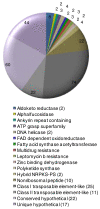
Results: The total assembly was 41.95 Mbp and contains 11,799 gene models of 50 amino acids or more. Comparison against two sequenced BACs showed that complex regions with a high GC content assembled effectively. Electrophoretic karyotyping showed distinct chromosomal polymorphisms between isolates 0-1 and 15A, and cytological karyotyping confirmed the presence of at least nine chromosomes. The genetic map spans 2477.7 cM and is composed of 243 markers in 25 linkage groups, and incorporates simple sequence repeat markers developed from the assembly. Among predicted genes, non-ribosomal peptide synthetases and efflux pumps in particular appear to have undergone a P. teres f. teres-specific expansion of non-orthologous gene families.
Conclusions: This study demonstrates that paired-end Solexa sequencing can successfully capture coding regions of a filamentous fungal genome. The assembly contains a plethora of predicted genes that have been implicated in a necrotrophic lifestyle and pathogenicity and presents a significant resource for examining the bases for P. teres f. teres pathogenicity.
© 2010 Ellwood et al.; licensee BioMed Central Ltd.
Figures




References
-
- Mathre DE. Compendium of Barley Diseases. 2. St Paul MN, American Phytopathological Society; 1997.
-
- Murray GM, Brennan JP. Estimating disease losses to the Australian barley industry. Aust Plant Pathol. 2010;39:85–96.
-
- Smedegård-Petersen V. Pyrenophora teres f. maculata f. nov. and Pyrenophora teres f. teres on barley in Denmark. Kgl Vet Landbohojsk Arsskr. 1971. pp. 124–144.
-
- Rau D, Attene G, Brown A, Nanni L, Maier F, Balmas V, Saba E, Schäfer W, Papa R. Phylogeny and evolution of mating-type genes from Pyrenophora teres, the causal agent of barley 'net blotch' disease. Curr Genet. 2007;51:377–392. - PubMed
-
- Campbell GF, Lucas JA, Crous PW. Evidence of recombination between net- and spot-type populations of Pyrenophora teres as determined by RAPD analysis. Mycol Res. 2002;106:602–608.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

