The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration
- PMID: 21068063
- PMCID: PMC2990206
- DOI: 10.1242/dev.052506
The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration
Abstract
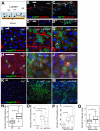
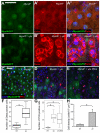
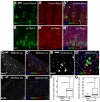
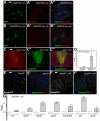
Intestinal stem cells (ISCs) in the adult Drosophila midgut proliferate to self-renew and to produce differentiating daughter cells that replace those lost as part of normal gut function. Intestinal stress induces the activation of Upd/Jak/Stat signalling, which promotes intestinal regeneration by inducing rapid stem cell proliferation. We have investigated the role of the Hippo (Hpo) pathway in the Drosophila intestine (midgut). Hpo pathway inactivation in either the ISCs or the differentiated enterocytes induces a phenotype similar to that observed under stress situations, including increased stem cell proliferation and expression of Jak/Stat pathway ligands. Hpo pathway targets are induced by stresses such as bacterial infection, suggesting that the Hpo pathway functions as a sensor of cellular stress in the differentiated cells of the midgut. In addition, Yki, the pro-growth transcription factor target of the Hpo pathway, is required in ISCs to drive the proliferative response to stress. Our results suggest that the Hpo pathway is a mediator of the regenerative response in the Drosophila midgut.
Figures



References
-
- Agaisse H., Petersen U. M., Boutros M., Mathey-Prevot B., Perrimon N. (2003). Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev. Cell 5, 441-450 - PubMed
-
- Arbouzova N. I., Zeidler M. P. (2006). JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development 133, 2605-2616 - PubMed
-
- Bach E. A., Ekas L. A., Ayala-Camargo A., Flaherty M. S., Lee H., Perrimon N., Baeg G. H. (2007). GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr. Patterns 7, 323-331 - PubMed
-
- Bando T., Mito T., Maeda Y., Nakamura T., Ito F., Watanabe T., Ohuchi H., Noji S. (2009). Regulation of leg size and shape by the Dachsous/Fat signalling pathway during regeneration. Development 136, 2235-2245 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

