Origin of the 1918 pandemic H1N1 influenza A virus as studied by codon usage patterns and phylogenetic analysis
- PMID: 21068184
- PMCID: PMC3004067
- DOI: 10.1261/rna.2395211
Origin of the 1918 pandemic H1N1 influenza A virus as studied by codon usage patterns and phylogenetic analysis
Abstract
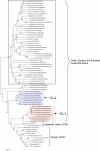
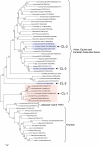
The pandemic of 1918 was caused by an H1N1 influenza A virus, which is a negative strand RNA virus; however, little is known about the nature of its direct ancestral strains. Here we applied a broad genetic and phylogenetic analysis of a wide range of influenza virus genes, in particular the PB1 gene, to gain information about the phylogenetic relatedness of the 1918 H1N1 virus. We compared the RNA genome of the 1918 strain to many other influenza strains of different origin by several means, including relative synonymous codon usage (RSCU), effective number of codons (ENC), and phylogenetic relationship. We found that the PB1 gene of the 1918 pandemic virus had ENC values similar to the H1N1 classical swine and human viruses, but different ENC values from avian as well as H2N2 and H3N2 human viruses. Also, according to the RSCU of the PB1 gene, the 1918 virus grouped with all human isolates and "classical" swine H1N1 viruses. The phylogenetic studies of all eight RNA gene segments of influenza A viruses may indicate that the 1918 pandemic strain originated from a H1N1 swine virus, which itself might be derived from a H1N1 avian precursor, which was separated from the bulk of other avian viruses in toto a long time ago. The high stability of the RSCU pattern of the PB1 gene indicated that the integrity of RNA structure is more important for influenza virus evolution than previously thought.
Figures


References
-
- Antonovics J, Hood ME, Baker CH 2006. Molecular virology: Was the 1918 flu avian in origin? Nature 440: E9 doi: 10.1038/nature04824 - PubMed
-
- Chun J 1919. Influenza including its infection among pigs. Nat Med J China (Peking) 5: 34–44
-
- Cortazzo P, Cervenansky C, Marin M, Reiss C, Ehrlich R, Deana A 2002. Silent mutations affect in vivo protein folding in Escherichia coli. Biochem Biophys Res Commun 293: 537–541 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
