The exozyme model: a continuum of functionally distinct complexes
- PMID: 21068185
- PMCID: PMC3004051
- DOI: 10.1261/rna.2364811
The exozyme model: a continuum of functionally distinct complexes
Abstract
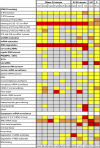
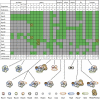
Exosome complexes are composed of 10 to 11 subunits and are involved in multiple facets of 3' → 5' RNA processing and turnover. The current paradigm stipulates that a uniform, stoichiometric core exosome, composed of single copies of each subunit, carries out all RNA metabolic functions in vivo. While core composition is well established in vitro, available genetic, cell biological, proteomic, and transcriptomic data raise questions about whether individual subunits contribute to RNA metabolic functions exclusively within the complex. Here, we recount the current understanding of the core exosome model and show predictions of the core model that are not satisfied by the available evidence. To resolve this discrepancy, we propose the exozyme hypothesis, a novel model stipulating that while exosome subunits can and do carry out certain functions within the core, subsets of exosome subunits and cofactors also assemble into a continuum of compositionally distinct complexes--exozymes--with different RNA specificities. The exozyme model is consistent with all published data and provides a new framework for understanding the general mechanisms and regulation of RNA processing and turnover.
Figures

References
-
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21: 87–96 - PubMed
-
- Allmang C, Tollervey D 1998. The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J Mol Biol 278: 67–78 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
