A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry
- PMID: 21068237
- PMCID: PMC3020023
- DOI: 10.1128/JVI.02062-10
A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry
Abstract
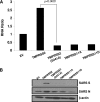
Spike (S) proteins, the defining projections of the enveloped coronaviruses (CoVs), mediate cell entry by connecting viruses to plasma membrane receptors and by catalyzing subsequent virus-cell membrane fusions. The latter membrane fusion requires an S protein conformational flexibility that is facilitated by proteolytic cleavages. We hypothesized that the most relevant cellular proteases in this process are those closely linked to host cell receptors. The primary receptor for the human severe acute respiratory syndrome CoV (SARS) CoV is angiotensin-converting enzyme 2 (ACE2). ACE2 immunoprecipitation captured transmembrane protease/serine subfamily member 2 (TMPRSS2), a known human airway and alveolar protease. ACE2 and TMPRSS2 colocalized on cell surfaces and enhanced the cell entry of both SARS S-pseudotyped HIV and authentic SARS-CoV. Enhanced entry correlated with TMPRSS2-mediated proteolysis of both S and ACE2. These findings indicate that a cell surface complex comprising a primary receptor and a separate endoprotease operates as a portal for activation of SARS-CoV cell entry.
Figures




References
-
- Afar, D. E., I. Vivanco, R. S. Hubert, J. Kuo, E. Chen, D. C. Saffran, A. B. Raitano, and A. Jakobovits. 2001. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 61:1686-1692. - PubMed
-
- Arpin, N., and P. J. Talbot. 1990. Molecular characterization of the 229E strain of human coronavirus. Adv. Exp. Med. Biol. 276:73-80. - PubMed
-
- Beaufort, N., D. Leduc, H. Eguchi, K. Mengele, D. Hellmann, T. Masegi, T. Kamimura, S. Yasuoka, F. Fend, M. Chignard, and D. Pidard. 2007. The human airway trypsin-like protease modulates the urokinase receptor (uPAR, CD87) structure and functions. Am. J. Physiol. Lung Cell. Mol. Physiol. 292:L1263-L1272. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

