Herpes simplex virus type 1 helicase-primase: DNA binding and consequent protein oligomerization and primase activation
- PMID: 21068246
- PMCID: PMC3019990
- DOI: 10.1128/JVI.01690-10
Herpes simplex virus type 1 helicase-primase: DNA binding and consequent protein oligomerization and primase activation
Abstract
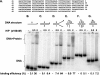
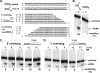
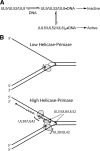
The heterotrimeric helicase-primase complex of herpes simplex virus type I (HSV-1), consisting of UL5, UL8, and UL52, possesses 5' to 3' helicase, single-stranded DNA (ssDNA)-dependent ATPase, primase, and DNA binding activities. In this study we confirm that the UL5-UL8-UL52 complex has higher affinity for forked DNA than for ssDNA and fails to bind to fully annealed double-stranded DNA substrates. In addition, we show that a single-stranded overhang of greater than 6 nucleotides is required for efficient enzyme loading and unwinding. Electrophoretic mobility shift assays and surface plasmon resonance analysis provide additional quantitative information about how the UL5-UL8-UL52 complex associates with the replication fork. Although it has previously been reported that in the absence of DNA and nucleoside triphosphates the UL5-UL8-UL52 complex exists as a monomer in solution, we now present evidence that in the presence of forked DNA and AMP-PNP, higher-order complexes can form. Electrophoretic mobility shift assays reveal two discrete complexes with different mobilities only when helicase-primase is bound to DNA containing a single-stranded region, and surface plasmon resonance analysis confirms larger amounts of the complex bound to forked substrates than to single-overhang substrates. Furthermore, we show that primase activity exhibits a cooperative dependence on protein concentration while ATPase and helicase activities do not. Taken together, these data suggest that the primase activity of the helicase-primase requires formation of a dimer or higher-order structure while ATPase activity does not. Importantly, this provides a simple mechanism for generating a two-polymerase replisome at the replication fork.
Figures







References
-
- Aslani, A., M. Olsson, and P. Elias. 2002. ATP-dependent unwinding of a minimal origin of DNA replication by the origin-binding protein and the single-strand DNA-binding protein ICP8 from herpes simplex virus type I. J. Biol. Chem. 277:41204-41212. - PubMed
-
- Biswas, N., and S. K. Weller. 1999. A mutation in the C-terminal putative Zn2+ finger motif of UL52 severely affects the biochemical activities of the HSV-1 helicase-primase subcomplex. J. Biol. Chem. 274:8068-8076. - PubMed
-
- Biswas, N., and S. K. Weller. 2001. The UL5 and UL52 subunits of the herpes simplex virus type 1 helicase-primase subcomplex exhibit a complex interdependence for DNA binding. J. Biol. Chem. 276:17610-17619. - PubMed
-
- Biswas, S., and H. J. Field. 2008. Herpes simplex virus helicase-primase inhibitors: recent findings from the study of drug resistance mutations. Antivir. Chem. Chemother. 19:1-6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

