Cleavage at the 586 amino acid caspase-6 site in mutant huntingtin influences caspase-6 activation in vivo
- PMID: 21068307
- PMCID: PMC3074336
- DOI: 10.1523/JNEUROSCI.2071-10.2010
Cleavage at the 586 amino acid caspase-6 site in mutant huntingtin influences caspase-6 activation in vivo
Abstract
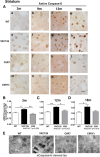
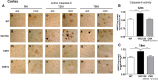
Caspase cleavage of huntingtin (htt) and nuclear htt accumulation represent early neuropathological changes in brains of patients with Huntington's disease (HD). However, the relationship between caspase cleavage of htt and caspase activation patterns in the pathogenesis of HD remains poorly understood. The lack of a phenotype in YAC mice expressing caspase-6-resistant (C6R) mutant htt (mhtt) highlights proteolysis of htt at the 586 aa caspase-6 (casp6) site as a key mechanism in the pathology of HD. The goal of this study was to investigate how proteolysis of htt at residue 586 plays a role in the pathogenesis of HD and determine whether inhibiting casp6 cleavage of mhtt alters cell-death pathways in vivo. Here we demonstrate that activation of casp6, and not caspase-3, is observed before onset of motor abnormalities in human and murine HD brain. Active casp6 levels correlate directly with CAG size and inversely with age of onset. In contrast, in vivo expression of C6R mhtt attenuates caspase activation. Increased casp6 activity and apoptotic cell death is evident in primary striatal neurons expressing caspase-cleavable, but not C6R, mhtt after NMDA application. Pretreatment with a casp6 inhibitor rescues the apoptotic cell death observed in this paradigm. These data demonstrate that activation of casp6 is an early marker of disease in HD. Furthermore, these data provide a clear link between excitotoxic pathways and proteolysis and suggest that C6R mhtt protects against neurodegeneration by influencing the activation of neuronal cell-death and excitotoxic pathways operative in HD.
Figures




References
-
- Allsopp TE, McLuckie J, Kerr LE, Macleod M, Sharkey J, Kelly JS. Caspase 6 activity initiates caspase 3 activation in cerebellar granule cell apoptosis. Cell Death Differ. 2000;7:984–993. - PubMed
-
- Andrew SE, Goldberg YP, Theilmann J, Zeisler J, Hayden MR. A CCG repeat polymorphism adjacent to the CAG repeat in the Huntington disease gene: implications for diagnostic accuracy and predictive testing. Hum Mol Genet. 1994;3:65–67. - PubMed
-
- Aziz NA, van der Burg JM, Landwehrmeyer GB, Brundin P, Stijnen T, Roos RA EHDI Study Group. Weight loss in Huntington disease increases with higher CAG repeat number. Neurology. 2008;71:1506–1513. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
