RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1
- PMID: 21068368
- PMCID: PMC2996456
- DOI: 10.1073/pnas.1009231107
RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1
Abstract
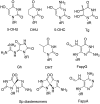
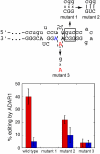
Editing of the pre-mRNA for the DNA repair enzyme NEIL1 causes a lysine to arginine change in the lesion recognition loop of the protein. The two forms of NEIL1 are shown here to have distinct enzymatic properties. The edited form removes thymine glycol from duplex DNA 30 times more slowly than the form encoded in the genome, whereas editing enhances repair of the guanidinohydantoin lesion by NEIL1. In addition, we show that the NEIL1 recoding site is a preferred editing site for the RNA editing adenosine deaminase ADAR1. The edited adenosine resides in an A-C mismatch in a hairpin stem formed by pairing of exon 6 to the immediate upstream intron 5 sequence. As expected for an ADAR1 site, editing at this position is increased in human cells treated with interferon α. These results suggest a unique regulatory mechanism for DNA repair and extend our understanding of the impact of RNA editing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Grosjean H, Benne R. Modification and editing of RNA. Washington, D.C.: ASM Press; 1998.
-
- Higuchi M, et al. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. - PubMed
-
- Burns CM, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387(6630):303–308. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

