Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae
- PMID: 21068384
- PMCID: PMC3013015
- DOI: 10.1074/jbc.M110.188805
Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae
Abstract
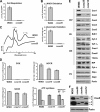
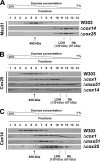
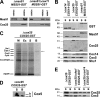
In the yeast Saccharomyces cerevisiae, mitochondrial cytochrome c oxidase (COX) biogenesis is translationally regulated. Mss51, a specific COX1 mRNA translational activator and Cox1 chaperone, drives the regulatory mechanism. During translation and post-translationally, newly synthesized Cox1 physically interacts with a complex of proteins involving Ssc1, Mss51, and Cox14, which eventually hand over Cox1 to the assembly pathway. This step is probably catalyzed by assembly chaperones such as Shy1 in a process coupled to the release of Ssc1-Mss51 from the complex. Impaired COX assembly results in the trapping of Mss51 in the complex, thus limiting its availability for COX1 mRNA translation. An exception is a null mutation in COX14 that does not affect Cox1 synthesis because the Mss51 trapping complexes become unstable, and Mss51 is readily available for translation. Here we present evidence showing that Cox25 is a new essential COX assembly factor that plays some roles similar to Cox14. A null mutation in COX25 by itself or in combination with other COX mutations does not affect Cox1 synthesis. Cox25 is an inner mitochondrial membrane intrinsic protein with a hydrophilic C terminus protruding into the matrix. Cox25 is an essential component of the complexes containing newly synthesized Cox1, Ssc1, Mss51, and Cox14. In addition, Cox25 is also found to interact with Shy1 and Cox5 in a complex that does not contain Mss51. These results suggest that once Ssc1-Mss51 are released from the Cox1 stabilization complex, Cox25 continues to interact with Cox14 and Cox1 to facilitate the formation of multisubunit COX assembly intermediates.
Figures
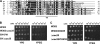


Similar articles
-
Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis.Mol Cell Biol. 2010 Jan;30(1):245-59. doi: 10.1128/MCB.00983-09. Mol Cell Biol. 2010. PMID: 19858289 Free PMC article.
-
Dual functions of Mss51 couple synthesis of Cox1 to assembly of cytochrome c oxidase in Saccharomyces cerevisiae mitochondria.Mol Biol Cell. 2009 Oct;20(20):4371-80. doi: 10.1091/mbc.e09-06-0522. Epub 2009 Aug 26. Mol Biol Cell. 2009. PMID: 19710419 Free PMC article.
-
Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria.J Cell Biol. 2010 Oct 4;191(1):141-54. doi: 10.1083/jcb.201007026. Epub 2010 Sep 27. J Cell Biol. 2010. PMID: 20876281 Free PMC article.
-
Biogenesis of cytochrome oxidase-sophisticated assembly lines in the mitochondrial inner membrane.Gene. 2005 Jul 18;354:43-52. doi: 10.1016/j.gene.2005.03.017. Gene. 2005. PMID: 15905047 Review.
-
Cytochrome c oxidase biogenesis - from translation to early assembly of the core subunit COX1.FEBS Lett. 2023 Jun;597(12):1569-1578. doi: 10.1002/1873-3468.14671. Epub 2023 May 31. FEBS Lett. 2023. PMID: 37247261 Review.
Cited by
-
Defects in mitochondrial fatty acid synthesis result in failure of multiple aspects of mitochondrial biogenesis in Saccharomyces cerevisiae.Mol Microbiol. 2013 Nov;90(4):824-40. doi: 10.1111/mmi.12402. Epub 2013 Oct 10. Mol Microbiol. 2013. PMID: 24102902 Free PMC article.
-
Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core.Biochim Biophys Acta. 2012 Jun;1817(6):883-97. doi: 10.1016/j.bbabio.2011.09.005. Epub 2011 Sep 16. Biochim Biophys Acta. 2012. PMID: 21958598 Free PMC article. Review.
-
Cox2p of yeast cytochrome oxidase assembles as a stand-alone subunit with the Cox1p and Cox3p modules.J Biol Chem. 2018 Oct 26;293(43):16899-16911. doi: 10.1074/jbc.RA118.004138. Epub 2018 Sep 17. J Biol Chem. 2018. PMID: 30224355 Free PMC article.
-
The Complexity of Mitochondrial Complex IV: An Update of Cytochrome c Oxidase Biogenesis in Plants.Int J Mol Sci. 2018 Feb 27;19(3):662. doi: 10.3390/ijms19030662. Int J Mol Sci. 2018. PMID: 29495437 Free PMC article. Review.
-
Oms1 associates with cytochrome c oxidase assembly intermediates to stabilize newly synthesized Cox1.Mol Biol Cell. 2016 May 15;27(10):1570-80. doi: 10.1091/mbc.E15-12-0811. Epub 2016 Mar 30. Mol Biol Cell. 2016. PMID: 27030670 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

