Aspartate 141 is the fourth ligand of the oxygen-sensing [4Fe-4S]2+ cluster of Bacillus subtilis transcriptional regulator Fnr
- PMID: 21068385
- PMCID: PMC3023498
- DOI: 10.1074/jbc.M110.191940
Aspartate 141 is the fourth ligand of the oxygen-sensing [4Fe-4S]2+ cluster of Bacillus subtilis transcriptional regulator Fnr
Abstract
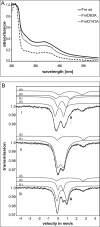
The Bacillus subtilis redox regulator Fnr controls genes of the anaerobic metabolism in response to low oxygen tension. An unusual structure for the oxygen-sensing [4Fe-4S](2+) cluster was detected by a combination of genetic experiments with UV-visible and Mössbauer spectroscopy. Asp-141 was identified as the fourth iron-sulfur cluster ligand besides three Cys residues. Exchange of Asp-141 with Ala abolished functional in vivo complementation of an fnr knock-out strain by the mutagenized fnr gene and in vitro DNA binding of the recombinant regulator FnrD141A. In contrast, substitution of Asp-141 with Cys preserved [4Fe-4S](2+) structure and regulator function.
Figures

Similar articles
-
Bacillus subtilis Fnr senses oxygen via a [4Fe-4S] cluster coordinated by three cysteine residues without change in the oligomeric state.Mol Microbiol. 2006 Jun;60(6):1432-45. doi: 10.1111/j.1365-2958.2006.05198.x. Mol Microbiol. 2006. PMID: 16796679
-
Substitution of leucine 28 with histidine in the Escherichia coli transcription factor FNR results in increased stability of the [4Fe-4S](2+) cluster to oxygen.J Biol Chem. 2000 Mar 3;275(9):6234-40. doi: 10.1074/jbc.275.9.6234. J Biol Chem. 2000. PMID: 10692418
-
Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster.FEMS Microbiol Rev. 1998 Dec;22(5):341-52. doi: 10.1111/j.1574-6976.1998.tb00375.x. FEMS Microbiol Rev. 1998. PMID: 9990723 Review.
-
Anaerobic transcription activation in Bacillus subtilis: identification of distinct FNR-dependent and -independent regulatory mechanisms.EMBO J. 1995 Dec 1;14(23):5984-94. doi: 10.1002/j.1460-2075.1995.tb00287.x. EMBO J. 1995. PMID: 8846791 Free PMC article.
-
Reactions of nitric oxide and oxygen with the regulator of fumarate and nitrate reduction, a global transcriptional regulator, during anaerobic growth of Escherichia coli.Methods Enzymol. 2008;437:191-209. doi: 10.1016/S0076-6879(07)37011-0. Methods Enzymol. 2008. PMID: 18433630 Review.
Cited by
-
NsrR from Streptomyces coelicolor is a nitric oxide-sensing [4Fe-4S] cluster protein with a specialized regulatory function.J Biol Chem. 2015 May 15;290(20):12689-704. doi: 10.1074/jbc.M115.643072. Epub 2015 Mar 14. J Biol Chem. 2015. PMID: 25771538 Free PMC article.
-
Novel, oxygen-insensitive group 5 [NiFe]-hydrogenase in Ralstonia eutropha.Appl Environ Microbiol. 2013 Sep;79(17):5137-45. doi: 10.1128/AEM.01576-13. Epub 2013 Jun 21. Appl Environ Microbiol. 2013. PMID: 23793632 Free PMC article.
-
Characterization of auxiliary iron-sulfur clusters in a radical S-adenosylmethionine enzyme PqqE from Methylobacterium extorquens AM1.FEBS Open Bio. 2017 Oct 18;7(12):1864-1879. doi: 10.1002/2211-5463.12314. eCollection 2017 Dec. FEBS Open Bio. 2017. PMID: 29226074 Free PMC article.
-
Reassessing the Structure and Function Relationship of the O2 Sensing Transcription Factor FNR.Antioxid Redox Signal. 2018 Dec 20;29(18):1830-1840. doi: 10.1089/ars.2017.7365. Epub 2017 Nov 14. Antioxid Redox Signal. 2018. PMID: 28990402 Free PMC article. Review.
-
The rational design of iron-sulfur cluster binding site for prolonged stability in magnetoreceptor MagR.Front Mol Biosci. 2022 Nov 10;9:1051943. doi: 10.3389/fmolb.2022.1051943. eCollection 2022. Front Mol Biosci. 2022. PMID: 36438652 Free PMC article.
References
-
- Green J., Crack J. C., Thomson A. J., LeBrun N. E. (2009) Curr. Opin. Microbiol. 12, 145–151 - PubMed
-
- Kiley P. J., Beinert H. (2003) Curr. Opin. Microbiol. 6, 181–185 - PubMed
-
- Reents H., Gruner I., Harmening U., Böttger L. H., Layer G., Heathcote P., Trautwein A. X., Jahn D., Härtig E. (2006) Mol. Microbiol. 60, 1432–1445 - PubMed
-
- Britt R. D., Sauer K., Klein M. P., Knaff D. B., Kriauciunas A., Yu C. A., Yu L., Malkin R. (1991) Biochemistry 30, 1892–1901 - PubMed
-
- Lovenberg W., Buchanan B. B., Rabinowitz J. C. (1963) J. Biol. Chem. 238, 3899–3913 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

