Global genome analysis of the shikimic acid pathway reveals greater gene loss in host-associated than in free-living bacteria
- PMID: 21070645
- PMCID: PMC3018139
- DOI: 10.1186/1471-2164-11-628
Global genome analysis of the shikimic acid pathway reveals greater gene loss in host-associated than in free-living bacteria
Abstract
Background: A central tenet in biochemistry for over 50 years has held that microorganisms, plants and, more recently, certain apicomplexan parasites synthesize essential aromatic compounds via elaboration of a complete shikimic acid pathway, whereas metazoans lacking this pathway require a dietary source of these compounds. The large number of sequenced bacterial and archaean genomes now available for comparative genomic analyses allows the fundamentals of this contention to be tested in prokaryotes. Using Hidden Markov Model profiles (HMM profiles) to identify all known enzymes of the pathway, we report the presence of genes encoding shikimate pathway enzymes in the hypothetical proteomes constructed from the genomes of 488 sequenced prokaryotes.
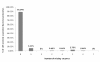
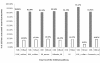
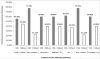
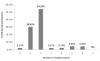
Results: Amongst free-living prokaryotes most Bacteria possess, as expected, genes encoding a complete shikimic acid pathway, whereas of the culturable Archaea, only one was found to have a complete complement of recognisable enzymes in its predicted proteome. It may be that in the Archaea, the primary amino-acid sequences of enzymes of the pathway are highly divergent and so are not detected by HMM profiles. Alternatively, structurally unrelated (non-orthologous) proteins might be performing the same biochemical functions as those encoding recognized genes of the shikimate pathway. Most surprisingly, 30% of host-associated (mutualistic, commensal and pathogenic) bacteria likewise do not possess a complete shikimic acid pathway. Many of these microbes show some degree of genome reduction, suggesting that these host-associated bacteria might sequester essential aromatic compounds from a parasitised host, as a 'shared metabolic adaptation' in mutualistic symbiosis, or obtain them from other consorts having the complete biosynthetic pathway. The HMM results gave 84% agreement when compared against data in the highly curated BioCyc reference database of genomes and metabolic pathways.
Conclusions: These results challenge the conventional belief that the shikimic acid pathway is universal and essential in prokaryotes. The possibilities that non-orthologous enzymes catalyse reactions in this pathway (especially in the Archaea), or that there exist specific uptake mechanisms for the acquisition of shikimate intermediates or essential pathway products, warrant further examination to better understand the precise metabolic attributes of host-beneficial and pathogenic bacteria.
Figures
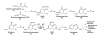

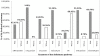
Similar articles
-
Genome-wide identification, domain architectures and phylogenetic analysis provide new insights into the early evolution of shikimate pathway in prokaryotes.Mol Phylogenet Evol. 2014 Jun;75:154-64. doi: 10.1016/j.ympev.2014.02.015. Epub 2014 Mar 3. Mol Phylogenet Evol. 2014. PMID: 24602988
-
Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements.Eukaryot Cell. 2006 Sep;5(9):1517-31. doi: 10.1128/EC.00106-06. Eukaryot Cell. 2006. PMID: 16963634 Free PMC article.
-
Enzymes of the shikimic acid pathway encoded in the genome of a basal metazoan, Nematostella vectensis, have microbial origins.Proc Natl Acad Sci U S A. 2008 Feb 19;105(7):2533-7. doi: 10.1073/pnas.0707388105. Epub 2008 Feb 11. Proc Natl Acad Sci U S A. 2008. PMID: 18268342 Free PMC article.
-
The shikimate pathway: review of amino acid sequence, function and three-dimensional structures of the enzymes.Crit Rev Microbiol. 2015 Jun;41(2):172-89. doi: 10.3109/1040841X.2013.813901. Epub 2013 Aug 6. Crit Rev Microbiol. 2015. PMID: 23919299 Review.
-
Metabolic Engineering of Shikimic Acid Biosynthesis Pathway for the Production of Shikimic Acid and Its Branched Products in Microorganisms: Advances and Prospects.Molecules. 2022 Jul 26;27(15):4779. doi: 10.3390/molecules27154779. Molecules. 2022. PMID: 35897952 Free PMC article. Review.
Cited by
-
Minimum Inhibitory Concentration of Glyphosate and a Glyphosate-Containing Herbicide in Salmonella enterica Isolates Originating from Different Time Periods, Hosts, and Serovars.Eur J Microbiol Immunol (Bp). 2019 May 20;9(2):35-41. doi: 10.1556/1886.2019.00005. eCollection 2019 Jun 3. Eur J Microbiol Immunol (Bp). 2019. PMID: 31223494 Free PMC article.
-
Identification of Volatiles of the Dinoflagellate Prorocentrum cordatum.Mar Drugs. 2022 May 30;20(6):371. doi: 10.3390/md20060371. Mar Drugs. 2022. PMID: 35736174 Free PMC article.
-
Uncultivated DPANN archaea are ubiquitous inhabitants of global oxygen deficient zones with diverse metabolic potential.bioRxiv [Preprint]. 2023 Oct 30:2023.10.30.564641. doi: 10.1101/2023.10.30.564641. bioRxiv. 2023. Update in: mBio. 2024 Mar 13;15(3):e0291823. doi: 10.1128/mbio.02918-23. PMID: 37961710 Free PMC article. Updated. Preprint.
-
Computational modelling provides insight into the effects of glyphosate on the shikimate pathway in the human gut microbiome.Curr Res Toxicol. 2020 Apr 22;1:25-33. doi: 10.1016/j.crtox.2020.04.001. eCollection 2020 Jun 10. Curr Res Toxicol. 2020. PMID: 34345834 Free PMC article.
-
In vivo and in silico determination of essential genes of Campylobacter jejuni.BMC Genomics. 2011 Nov 1;12:535. doi: 10.1186/1471-2164-12-535. BMC Genomics. 2011. PMID: 22044676 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

