A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3
- PMID: 21071421
- PMCID: PMC3064776
- DOI: 10.1093/nar/gkq1118
A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3
Abstract
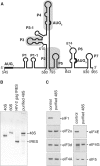
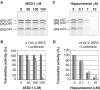
Translation initiation on HIV genomic RNA relies on both cap and Internal Ribosome Entry Site (IRES) dependant mechanisms that are regulated throughout the cell cycle. During a unique phenomenon, the virus recruits initiation complexes through RNA structures located within Gag coding sequence, downstream of the initiation codon. We analyzed initiation complexes paused on the HIV-2 gag IRES and revealed that they contain all the canonical initiation factors except eIF4E and eIF1. We report that eIF3 and the small ribosomal subunit bind HIV RNA within gag open reading frame. We thus propose a novel two step model whereby the initial event is the formation of a ternary eIF3/40S/IRES complex. In a second step, dependent on most of the canonical initiation factors, the complex is rearranged to transfer the ribosome on the initiation codons. The absolute requirement of this large structure for HIV translation defines a new function for a coding region. Moreover, the level of information compaction within this viral genome reveals an additional level of evolutionary constraint on the coding sequence. The conservation of this IRES and its properties in rapidly evolving viruses suggest an important role in the virus life cycle and highlight an attractive new therapeutic target.
Figures

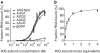
References
-
- Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:1–34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

