F2Dock: fast Fourier protein-protein docking
- PMID: 21071796
- PMCID: PMC3058388
- DOI: 10.1109/TCBB.2009.57
F2Dock: fast Fourier protein-protein docking
Abstract
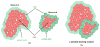
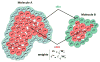
The functions of proteins are often realized through their mutual interactions. Determining a relative transformation for a pair of proteins and their conformations which form a stable complex, reproducible in nature, is known as docking. It is an important step in drug design, structure determination, and understanding function and structure relationships. In this paper, we extend our nonuniform fast Fourier transform-based docking algorithm to include an adaptive search phase (both translational and rotational) and thereby speed up its execution. We have also implemented a multithreaded version of the adaptive docking algorithm for even faster execution on multicore machines. We call this protein-protein docking code F2Dock (F2 = Fast Fourier). We have calibrated F2Dock based on an extensive experimental study on a list of benchmark complexes and conclude that F2Dock works very well in practice. Though all docking results reported in this paper use shape complementarity and Coulombic-potential-based scores only, F2Dock is structured to incorporate Lennard-Jones potential and reranking docking solutions based on desolvation energy .
Figures


References
-
- Fruton JS. Proteins, Enzymes, Genes: The Interplay of Chemistry and Biology. Yale University Press; 1999.
-
- Leach AR. Molecular Modelling: Principles and Applications. 2. Pearson Education EMA; 2001.
-
- Gabb HA, Jackson RM, Sternberg MJE. Modelling protein docking using shape complementarity, electrostatics and biochemical information. Journal of Molecular Biology. 1997 September;272(1):106–120. - PubMed
-
- Klotz IM. Ligand-receptor energetics: A guide for the perplexed. John Wiley and Sons, Inc; 1997.
-
- Castrillon-Candas J, Siddavanahalli V, Bajaj C. Nonequispaced fourier transforms for protein-protein docking. The University of Texas at Austin; Austin TX USA: Oct, 2005. ICES Report 05-44.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

