Evolutionary history of tissue kallikreins
- PMID: 21072173
- PMCID: PMC2967472
- DOI: 10.1371/journal.pone.0013781
Evolutionary history of tissue kallikreins
Abstract
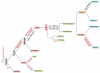
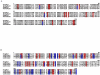
The gene family of human kallikrein-related peptidases (KLKs) encodes proteins with diverse and pleiotropic functions in normal physiology as well as in disease states. Currently, the most widely known KLK is KLK3 or prostate-specific antigen (PSA) that has applications in clinical diagnosis and monitoring of prostate cancer. The KLK gene family encompasses the largest contiguous cluster of serine proteases in humans which is not interrupted by non-KLK genes. This exceptional and unique characteristic of KLKs makes them ideal for evolutionary studies aiming to infer the direction and timing of gene duplication events. Previous studies on the evolution of KLKs were restricted to mammals and the emergence of KLKs was suggested about 150 million years ago (mya). In order to elucidate the evolutionary history of KLKs, we performed comprehensive phylogenetic analyses of KLK homologous proteins in multiple genomes including those that have been completed recently. Interestingly, we were able to identify novel reptilian, avian and amphibian KLK members which allowed us to trace the emergence of KLKs 330 mya. We suggest that a series of duplication and mutation events gave rise to the KLK gene family. The prominent feature of the KLK family is that it consists of tandemly and uninterruptedly arrayed genes in all species under investigation. The chromosomal co-localization in a single cluster distinguishes KLKs from trypsin and other trypsin-like proteases which are spread in different genetic loci. All the defining features of the KLKs were further found to be conserved in the novel KLK protein sequences. The study of this unique family will further assist in selecting new model organisms for functional studies of proteolytic pathways involving KLKs.
Conflict of interest statement
Figures




References
-
- Borgoño CA, Diamandis EP. The emerging roles of human tissue kallikreins in cancer. Nat Rev Cancer. 2004;4:876–890. - PubMed
-
- Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nat Rev Cancer. 2008;8:268–278. - PubMed
-
- Lundwall A, Band V, Blaber M, Clements JA, Courty Y, et al. A comprehensive nomenclature for serine proteases with homology to tissue kallikreins. Biol Chem. 2006;387:637–641. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

