New approaches toward recognition of nucleic acid triple helices
- PMID: 21073199
- PMCID: PMC3977315
- DOI: 10.1021/ar100113q
New approaches toward recognition of nucleic acid triple helices
Abstract
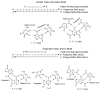
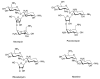
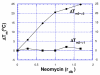
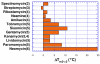
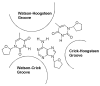
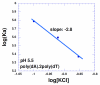
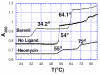
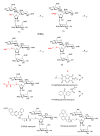
A DNA duplex can be recognized sequence-specifically in the major groove by an oligodeoxynucleotide (ODN). The resulting structure is a DNA triple helix, or triplex. The scientific community has invested significant research capital in the study of DNA triplexes because of their robust potential for providing new applications, including molecular biology tools and therapeutic agents. The triplex structures have inherent instabilities, however, and the recognition of DNA triplexes by small molecules has been attempted as a means of strengthening the three-stranded complex. Over the decades, the majority of work in the field has focused on heterocycles that intercalate between the triplex bases. In this Account, we present an alternate approach to recognition and stabilization of DNA triplexes. We show that groove recognition of nucleic acid triple helices can be achieved with aminosugars. Among these aminosugars, neomycin is the most effective aminoglycoside (groove binder) for stabilizing a DNA triple helix. It stabilizes both the TAT triplex and mixed-base DNA triplexes better than known DNA minor groove binders (which usually destabilize the triplex) and polyamines. Neomycin selectively stabilizes the triplex (TAT and mixed base) without any effect on the DNA duplex. The selectivity of neomycin likely originates from its potential and shape complementarity to the triplex Watson-Hoogsteen groove, making it the first molecule that selectively recognizes a triplex groove over a duplex groove. The groove recognition of aminoglycosides is not limited to DNA triplexes, but also extends to RNA and hybrid triple helical structures. Intercalator-neomycin conjugates are shown to simultaneously probe the base stacking and groove surface in the DNA triplex. Calorimetric and spectrosocopic studies allow the quantification of the effect of surface area of the intercalating moiety on binding to the triplex. These studies outline a novel approach to the recognition of DNA triplexes that incorporates the use of noncompeting binding sites. These principles of dual recognition should be applicable to the design of ligands that can bind any given nucleic acid target with nanomolar affinities and with high selectivity.
Figures






References
-
- Kool ET. Design of Triplex-forming Oligonucleotides for Binding DNA and RNA: Optimizing Affinity and Selectivity. New J. Chem. 1997;21:33–45.
-
- Dervan PB. Molecular Recognition of DNA by Small Molecules. Bioorg. Med. Chem. 2001;9:2215–2235. - PubMed
-
- Felsenfeld G, Davies D, Rich A. Formation of a Three-Stranded Polynucleotide Molecule. J. Amer. Chem. Soc. 1957;79:2023–2024.
-
- Felsenfeld G, Rich A. Studies on the Formation of Two and Three Stranded Polyribonucleotides. Biochim. Biophys. Acta. 1957;26:457–468. - PubMed
-
- Maher LJ, III, Wold B, Dervan PB. Inhibition of DNA Binding Proteins by Oligonucleotide-Directed Triple Helix Formation. Science. 1989;245:725–730. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

