The Vitis vinifera sugar transporter gene family: phylogenetic overview and macroarray expression profiling
- PMID: 21073695
- PMCID: PMC3095327
- DOI: 10.1186/1471-2229-10-245
The Vitis vinifera sugar transporter gene family: phylogenetic overview and macroarray expression profiling
Abstract
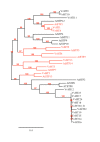
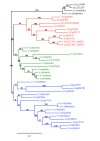
Background: In higher plants, sugars are not only nutrients but also important signal molecules. They are distributed through the plant via sugar transporters, which are involved not only in sugar long-distance transport via the loading and the unloading of the conducting complex, but also in sugar allocation into source and sink cells. The availability of the recently released grapevine genome sequence offers the opportunity to identify sucrose and monosaccharide transporter gene families in a woody species and to compare them with those of the herbaceous Arabidopsis thaliana using a phylogenetic analysis.
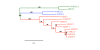
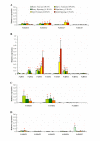
Results: In grapevine, one of the most economically important fruit crop in the world, it appeared that sucrose and monosaccharide transporter genes are present in 4 and 59 loci, respectively and that the monosaccharide transporter family can be divided into 7 subfamilies. Phylogenetic analysis of protein sequences has indicated that orthologs exist between Vitis and Arabidospis. A search for cis-regulatory elements in the promoter sequences of the most characterized transporter gene families (sucrose, hexoses and polyols transporters), has revealed that some of them might probably be regulated by sugars. To profile several genes simultaneously, we created a macroarray bearing cDNA fragments specific to 20 sugar transporter genes. This macroarray analysis has revealed that two hexose (VvHT1, VvHT3), one polyol (VvPMT5) and one sucrose (VvSUC27) transporter genes, are highly expressed in most vegetative organs. The expression of one hexose transporter (VvHT2) and two tonoplastic monosaccharide transporter (VvTMT1, VvTMT2) genes are regulated during berry development. Finally, three putative hexose transporter genes show a preferential organ specificity being highly expressed in seeds (VvHT3, VvHT5), in roots (VvHT2) or in mature leaves (VvHT5).
Conclusions: This study provides an exhaustive survey of sugar transporter genes in Vitis vinifera and revealed that sugar transporter gene families in this woody plant are strongly comparable to those of herbaceous species. Dedicated macroarrays have provided a Vitis sugar transporter genes expression profiling, which will likely contribute to understand their physiological functions in plant and berry development. The present results might also have a significant impact on our knowledge on plant sugar transporters.
Figures





References
-
- Zimmermann MH, Ziegler. In: Encyclopedia of Plant Physiology. Zimmermann MH, Milbum JA, editor. Berlin: Springer Verlag; 1975. List of sugars and sugar alcohols in sieve-tube exudates; pp. 480–503.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

