Impact of the Staphylococcus epidermidis LytSR two-component regulatory system on murein hydrolase activity, pyruvate utilization and global transcriptional profile
- PMID: 21073699
- PMCID: PMC2996381
- DOI: 10.1186/1471-2180-10-287
Impact of the Staphylococcus epidermidis LytSR two-component regulatory system on murein hydrolase activity, pyruvate utilization and global transcriptional profile
Abstract
Background: Staphylococcus epidermidis has emerged as one of the most important nosocomial pathogens, mainly because of its ability to colonize implanted biomaterials by forming a biofilm. Extensive studies are focused on the molecular mechanisms involved in biofilm formation. The LytSR two-component regulatory system regulates autolysis and biofilm formation in Staphylococcus aureus. However, the role of LytSR played in S. epidermidis remained unknown.
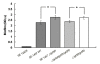
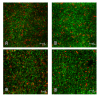
Results: In the present study, we demonstrated that lytSR knock-out in S. epidermidis did not alter susceptibility to Triton X-100 induced autolysis. Quantitative murein hydrolase assay indicated that disruption of lytSR in S. epidermidis resulted in decreased activities of extracellular murein hydrolases, although zymogram showed no apparent differences in murein hydrolase patterns between S. epidermidis strain 1457 and its lytSR mutant. Compared to the wild-type counterpart, 1457ΔlytSR produced slightly more biofilm, with significantly decreased dead cells inside. Microarray analysis showed that lytSR mutation affected the transcription of 164 genes (123 genes were upregulated and 41 genes were downregulated). Specifically, genes encoding proteins responsible for protein synthesis, energy metabolism were downregulated, while genes involved in amino acid and nucleotide biosynthesis, amino acid transporters were upregulated. Impaired ability to utilize pyruvate and reduced activity of arginine deiminase was observed in 1457ΔlytSR, which is consistent with the microarray data.
Conclusions: The preliminary results suggest that in S. epidermidis LytSR two-component system regulates extracellular murein hydrolase activity, bacterial cell death and pyruvate utilization. Based on the microarray data, it appears that lytSR inactivation induces a stringent response. In addition, LytSR may indirectly enhance biofilm formation by altering the metabolic status of the bacteria.
Figures





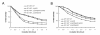
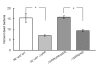

References
-
- Rupp ME, Archer GL. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin Infect Dis. 1994;19(2):231–243; quiz 244-235. - PubMed
-
- Bowden MG, Chen W, Singvall J, Xu Y, Peacock SJ, Valtulina V, Speziale P, Hook M. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology (Reading, England) 2005;151(Pt 5):1453–1464. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

