Metagenomic analysis of the turkey gut RNA virus community
- PMID: 21073719
- PMCID: PMC2991317
- DOI: 10.1186/1743-422X-7-313
Metagenomic analysis of the turkey gut RNA virus community
Abstract
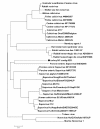
Viral enteric disease is an ongoing economic burden to poultry producers worldwide, and despite considerable research, no single virus has emerged as a likely causative agent and target for prevention and control efforts. Historically, electron microscopy has been used to identify suspect viruses, with many small, round viruses eluding classification based solely on morphology. National and regional surveys using molecular diagnostics have revealed that suspect viruses continuously circulate in United States poultry, with many viruses appearing concomitantly and in healthy birds. High-throughput nucleic acid pyrosequencing is a powerful diagnostic technology capable of determining the full genomic repertoire present in a complex environmental sample. We utilized the Roche/454 Life Sciences GS-FLX platform to compile an RNA virus metagenome from turkey flocks experiencing enteric disease. This approach yielded numerous sequences homologous to viruses in the BLAST nr protein database, many of which have not been described in turkeys. Our analysis of this turkey gut RNA metagenome focuses in particular on the turkey-origin members of the Picornavirales, the Caliciviridae, and the turkey Picobirnaviruses.
Figures

References
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

