Inhibition of core gene of HCV 3a genotype using synthetic and vector derived siRNAs
- PMID: 21073745
- PMCID: PMC2992066
- DOI: 10.1186/1743-422X-7-318
Inhibition of core gene of HCV 3a genotype using synthetic and vector derived siRNAs
Abstract
Background: Hepatitis C virus (HCV) is a major causative agent of liver associated diseases throughout the world, with genotype 3a responsible for most of the cases in Pakistan. Due to the limited efficiency of current therapy, RNA interference (RNAi) a novel regulatory and powerful silencing approach for molecular therapeutics through a sequence-specific RNA degradation process represents an alternative option.
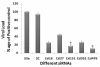
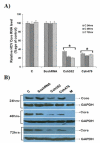
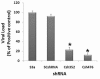
Results: The current study was purposed to assess and explore the possibility of RNAi to silence the HCV-3a Core gene expression, which play complex role in regulation of cell growth and host genes expression essential for infectivity and disease progression. To identify the potent siRNA target sites, 5 small interfering RNAs (siRNAs) against Core gene were designed and in vitro transcribed after consensus sequence analysis of different HCV-3a isolates. Antiviral effects of siRNAs showed upto 80% inhibition of Core gene expression by different siRNAs into Huh-7 cells as compared with Mock transfected and control siRNAs treated cells. For long lasting effect of siRNAs, vector based short hairpin siRNAs (shRNAs) were designed and tested against HCV-3a Core which resulted in a similar pattern of inhibition on RNA and protein expression of HCV Core as synthetic siRNAs. Furthermore, the efficacy of cell culture tested siRNA and shRNA, were evaluated for inhibition of HCV replication in HCV infected serum inoculated Huh-7 cells and a significant decrease in HCV viral copy number was observed.
Conclusions: Our results support the possibility of using consensus siRNA and shRNA-based molecular therapy as a promising strategy in effective inhibition of HCV-3a genotype.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

