Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues
- PMID: 21074044
- PMCID: PMC3032267
- DOI: 10.1016/j.cell.2010.10.008
Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues
Abstract
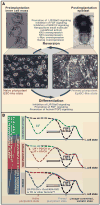
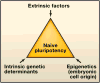
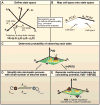
Direct reprogramming of somatic cells to induced pluripotent stem cells by ectopic expression of defined transcription factors has raised fundamental questions regarding the epigenetic stability of the differentiated cell state. In addition, evidence has accumulated that distinct states of pluripotency can interconvert through the modulation of both cell-intrinsic and exogenous factors. To fully realize the potential of in vitro reprogrammed cells, we need to understand the molecular and epigenetic determinants that convert one cell type into another. Here we review recent advances in this rapidly moving field and emphasize unresolved and controversial questions.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures



References
-
- Bloom JD, Meyer MM, Meinhold P, Otey CR, MacMillan D, Arnold FH. Evolving strategies for enzyme engineering. Curr Opin Struct Biol. 2005;15:447–452. - PubMed
-
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

