Hdac3 is essential for the maintenance of chromatin structure and genome stability
- PMID: 21075309
- PMCID: PMC3004468
- DOI: 10.1016/j.ccr.2010.10.022
Hdac3 is essential for the maintenance of chromatin structure and genome stability
Abstract
Hdac3 is essential for efficient DNA replication and DNA damage control. Deletion of Hdac3 impaired DNA repair and greatly reduced chromatin compaction and heterochromatin content. These defects corresponded to increases in histone H3K9,K14ac; H4K5ac; and H4K12ac in late S phase of the cell cycle, and histone deposition marks were retained in quiescent Hdac3-null cells. Liver-specific deletion of Hdac3 culminated in hepatocellular carcinoma. Whereas HDAC3 expression was downregulated in only a small number of human liver cancers, the mRNA levels of the HDAC3 cofactor NCOR1 were reduced in one-third of these cases. siRNA targeting of NCOR1 and SMRT (NCOR2) increased H4K5ac and caused DNA damage, indicating that the HDAC3/NCOR/SMRT axis is critical for maintaining chromatin structure and genomic stability.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
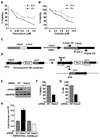


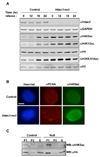

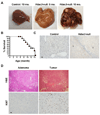
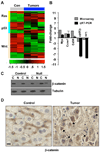

References
-
- Ashwell S, Zabludoff S. DNA damage detection and repair pathways--recent advances with inhibitors of checkpoint kinases in cancer therapy. Clin Cancer Res. 2008;14:4032–4037. - PubMed
-
- Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA077274/CA/NCI NIH HHS/United States
- P30CA68485/CA/NCI NIH HHS/United States
- T32 CA009582/CA/NCI NIH HHS/United States
- R01-CA77274/CA/NCI NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- 1F32CA138091-01/CA/NCI NIH HHS/United States
- R01 CA109355/CA/NCI NIH HHS/United States
- R01 CA141071/CA/NCI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- R01-CA109355/CA/NCI NIH HHS/United States
- F32 CA138091/CA/NCI NIH HHS/United States
- P30DK58404/DK/NIDDK NIH HHS/United States
- R01-CA64140/CA/NCI NIH HHS/United States
- R01 CA064140/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

