Mouse STAT2 restricts early dengue virus replication
- PMID: 21075352
- PMCID: PMC3310429
- DOI: 10.1016/j.chom.2010.10.007
Mouse STAT2 restricts early dengue virus replication
Abstract
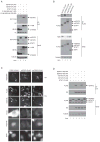
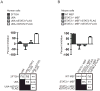
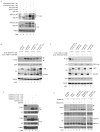
Dengue virus encodes several interferon antagonists. Among these the NS5 protein binds STAT2, a necessary component of the type I interferon signaling pathway, and targets it for degradation. We now demonstrate that the ability of dengue NS5 to associate with and degrade STAT2 is species specific. Thus, NS5 is able to bind and degrade human STAT2, but not mouse STAT2. This difference was exploited to demonstrate, absent manipulation of the viral genome, that NS5-mediated IFN antagonism is essential for efficient virus replication. Moreover, we demonstrate that differences in NS5 mediated binding and degradation between human and mouse STAT2 maps to a region within the STAT2 coiled-coil domain. By using STAT2(-/-) mice, we also demonstrate that mouse STAT2 restricts early dengue virus replication in vivo. These results suggest that overcoming this restriction through transgenic mouse technology may help in the development of a long-sought immune-competent mouse model of dengue virus infection.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures




References
-
- An J, Kimura-Kuroda J, Hirabayashi Y, Yasui K. Development of a novel mouse model for dengue virus infection. Virology. 1999;263:70–77. - PubMed
-
- Ashburn PM, Craig CF. Experimental investigations regarding the etiology of dengue fever. 1907. J Infect Dis. 2004;189:1747–1783. discussion 1744–1746. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

