Dengue virus-induced autophagy regulates lipid metabolism
- PMID: 21075353
- PMCID: PMC3026642
- DOI: 10.1016/j.chom.2010.10.006
Dengue virus-induced autophagy regulates lipid metabolism
Abstract
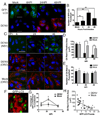
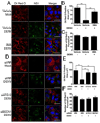
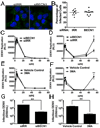
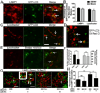
Autophagy influences numerous cellular processes, including innate and adaptive immunity against intracellular pathogens. However, some viruses, including dengue virus (DENV), usurp autophagy to enhance their replication. The mechanism for a positive role of autophagy in DENV infection is unclear. We present data that DENV induction of autophagy regulates cellular lipid metabolism. DENV infection leads to an autophagy-dependent processing of lipid droplets and triglycerides to release free fatty acids. This results in an increase in cellular β-oxidation, which generates ATP. These processes are required for efficient DENV replication. Importantly, exogenous fatty acids can supplant the requirement of autophagy in DENV replication. These results define a role for autophagy in DENV infection and provide a mechanism by which viruses can alter cellular lipid metabolism to promote their replication.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures


References
-
- Andre P, Perlemuter G, Budkowska A, Brechot C, Lotteau V. Hepatitis C virus particles and lipoprotein metabolism. Semin Liver Dis. 2005;25:93–104. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

