Epigenetic regulation of glial fibrillary acidic protein by DNA methylation in human malignant gliomas
- PMID: 21075782
- PMCID: PMC3018916
- DOI: 10.1093/neuonc/noq145
Epigenetic regulation of glial fibrillary acidic protein by DNA methylation in human malignant gliomas
Abstract
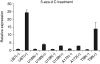
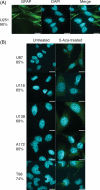
Glial fibrillary acidic protein (GFAP) is an intermediate filament expressed in glial cells that stabilizes and maintains the cytoskeleton of normal astrocytes. In glial tumors, GFAP expression is frequently lost with increasing grade of malignancy, suggesting that GFAP is important for maintaining glial cell morphology or regulating astrocytoma cell growth. Most permanent human glioma cell lines are GFAP negative by immunocytochemistry. Given that the GFAP gene is not mutated in human glioma specimens or glioma cell lines, we considered epigenetic mechanisms, such as promoter methylation, as a cause of silencing of GFAP in these tumors. In this study, we treated known GFAP-negative glioma cell lines with 5-aza-2'-deoxycytidine to examine GFAP promoter hypermethylation. Additionally, we performed bisulfite sequencing on primary glioma samples and glioma cell lines and showed an inverse relationship between GFAP promoter methylation status and GFAP expression. Using a gene reporter assay with the GFAP promoter cloned upstream of a luciferase gene, we showed that methylation of the GFAP promoter downregulates the expression of the luciferase gene. Our results suggest that epigenetic silencing of the GFAP gene through DNA methylation of its promoter region may be one mechanism by which GFAP is downregulated in human gliomas and glioma cell lines.
Figures
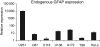



References
-
- Bansal K, Liang ML, Rutka JT. Molecular biology of human gliomas. Technol Cancer Res Treat. 2006;5:185–194. - PubMed
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Duffy PE, Huang YY, Rapport MM. The relationship of glial fibrillary acidic protein to the shape, motility, and differentiation of human astrocytoma cells. Exp Cell Res. 1982;139:145–157. - PubMed
-
- Duffy PE, Huang YY, Rapport MM, et al. Glial fibrillary acidic protein in giant cell tumors of brain and other gliomas. A possible relationship to malignancy, differentiation, and pleomorphism of glia. Acta Neuropathol. 1980;52:51–57. - PubMed
-
- Deck JH, Eng LF, Bigbee J, et al. The role of glial fibrillary acidic protein in the diagnosis of central nervous system tumors. Acta Neuropathol. 1978;42:183–190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

