Switch between life history strategies due to changes in glycolytic enzyme gene dosage in Saccharomyces cerevisiae
- PMID: 21075872
- PMCID: PMC3020566
- DOI: 10.1128/AEM.00808-10
Switch between life history strategies due to changes in glycolytic enzyme gene dosage in Saccharomyces cerevisiae
Abstract
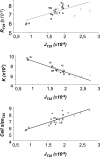
Adaptation is the process whereby a population or species becomes better fitted to its habitat through modifications of various life history traits which can be positively or negatively correlated. The molecular factors underlying these covariations remain to be elucidated. Using Saccharomyces cerevisiae as a model system, we have investigated the effects on life history traits of varying the dosage of genes involved in the transformation of resources into energy. Changing gene dosage for each of three glycolytic enzyme genes (hexokinase 2, phosphoglucose isomerase, and fructose-1,6-bisphosphate aldolase) resulted in variation in enzyme activities, glucose consumption rate, and life history traits (growth rate, carrying capacity, and cell size). However, the range of effects depended on which enzyme was expressed differently. Most interestingly, these changes revealed a genetic trade-off between carrying capacity and cell size, supporting the discovery of two extreme life history strategies already described in yeast populations: the "ants," which have lower glycolytic gene dosage, take up glucose slowly, and have a small cell size but reach a high carrying capacity, and the "grasshoppers," which have higher glycolytic gene dosage, consume glucose more rapidly, and allocate it to a larger cell size but reach a lower carrying capacity. These results demonstrate antagonist pleiotropy for glycolytic genes and show that altered dosage of a single gene drives a switch between two life history strategies in yeast.
Figures
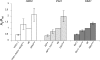

References
-
- Ahuatzi, D., P. Herrero, T. de la Cera, and F. Moreno. 2004. The glucose-regulated nuclear localization of hexokinase 2 in Saccharomyces cerevisiae is Mig1-dependent. J. Biol. Chem. 279:14440-14446. - PubMed
-
- Beatty, C. H., R. M. Bocek, and M. K. Young. 1975. Glycolytic control mechanisms in myometrium from pregnant rhesus monkeys. Biol. Reprod. 12:408-414. - PubMed
-
- Benevolensky, S. V., D. Clifton, and D. G. Fraenkel. 1994. The effect of increased phosphoglucose isomerase on glucose metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 269:4878-4882. - PubMed
-
- Bianconi, M. L. 2003. Calorimetric determination of thermodynamic parameters of reaction reveals different enthalpic compensations of the yeast hexokinase isozymes. J. Biol. Chem. 278:18709-18713. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

