Mutation and epigenetic molecular clocks in cancer
- PMID: 21076057
- PMCID: PMC3026845
- DOI: 10.1093/carcin/bgq239
Mutation and epigenetic molecular clocks in cancer
Abstract
A quandary of evolution is how to measure change over time. A natural inclination is to use morphologic criteria-the greater the differences between two phenotypes, the greater amount of time needed to evolve these differences. However, appearances may be deceiving, and another approach to infer time is with molecular clocks. Here, the greater the differences between two genomes, on average the greater the time since a common ancestor. Recent advances in DNA sequencing shed new light on how human cancers might evolve.
Figures
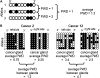
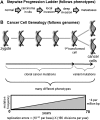

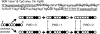

References
-
- Bromham L, et al. The modern molecular clock. Nat. Rev. Genet. 2003;4:216–224. - PubMed
-
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. - PubMed
-
- Tishkoff SA, et al. Implications of biogeography of human populations for ‘race’ and medicine. Nat. Genet. 2004;36:S21–S27. - PubMed
-
- Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

