The highly conserved eukaryotic DRG factors are required for efficient translation in a manner redundant with the putative RNA helicase Slh1
- PMID: 21076151
- PMCID: PMC3064805
- DOI: 10.1093/nar/gkq898
The highly conserved eukaryotic DRG factors are required for efficient translation in a manner redundant with the putative RNA helicase Slh1
Abstract
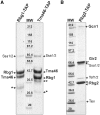
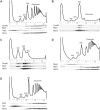
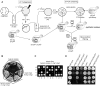
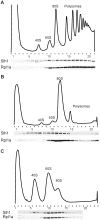
Eukaryotic and archaeal DRG factors are highly conserved proteins with characteristic GTPase motifs. This suggests their implication in a central biological process, which has so far escaped detection. We show here that the two Saccharomyces cerevisiae DRGs form distinct complexes, RBG1 and RBG2, and that the former co-fractionate with translating ribosomes. A genetic screen for triple synthetic interaction demonstrates that yeast DRGs have redundant function with Slh1, a putative RNA helicase also associating with translating ribosomes. Translation and cell growth are severely impaired in a triple mutant lacking both yeast DRGs and Slh1, but not in double mutants. This new genetic assay allowed us to characterize the roles of conserved motifs present in these proteins for efficient translation and/or association with ribosomes. Altogether, our results demonstrate for the first time a direct role of the highly conserved DRG factors in translation and indicate that this function is redundantly shared by three factors. Furthermore, our data suggest that important cellular processes are highly buffered against external perturbation and, consequently, that redundantly acting factors may escape detection in current high-throughput binary genetic interaction screens.
Figures

References
-
- Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002;317:41–72. - PubMed
-
- Caldon CE, March PE. Function of the universally conserved bacterial GTPases. Curr. Opin. Microbiol. 2003;6:135–139. - PubMed
-
- Li B, Trueb B. DRG represents a family of two closely related GTP-binding proteins. Biochim. Biophys. Acta. 2000;1491:196–204. - PubMed
-
- O'Connell A, Robin G, Kobe B, Botella JR. Biochemical characterization of Arabidopsis developmentally regulated G-proteins (DRGs) Protein Expr. Purif. 2009;67:88–95. - PubMed
-
- Sazuka T, Tomooka Y, Ikawa Y, Noda M, Kumar S. DRG: a novel developmentally regulated GTP-binding protein. Biochem. Biophys. Res. Commun. 1992;189:363–370. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

