The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases
- PMID: 21076411
- PMCID: PMC3003441
- DOI: 10.1038/ncb2121
The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases
Abstract
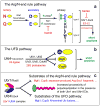
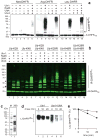
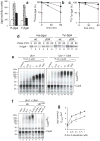
Substrates of the N-end rule pathway are recognized by the Ubr1 E3 ubiquitin ligase through their destabilizing amino-terminal residues. Our previous work showed that the Ubr1 E3 and the Ufd4 E3 together target an internal degradation signal (degron) of the Mgt1 DNA repair protein. Ufd4 is an E3 enzyme of the ubiquitin-fusion degradation (UFD) pathway that recognizes an N-terminal ubiquitin moiety. Here we show that the RING-type Ubr1 E3 and the HECT-type Ufd4 E3 interact, both physically and functionally. Although Ubr1 can recognize and polyubiquitylate an N-end rule substrate in the absence of Ufd4, the Ubr1-Ufd4 complex is more processive in that it produces a longer substrate-linked polyubiquitin chain. Conversely, Ubr1 can function as a polyubiquitylation-enhancing component of the Ubr1-Ufd4 complex in its targeting of UFD substrates. We also found that Ubr1 can recognize the N-terminal ubiquitin moiety. These and related advances unify two proteolytic systems that have been studied separately for two decades.
Conflict of interest statement
The authors declare that they have no competing financial interest.
Figures



Comment in
-
Working on a chain: E3s ganging up for ubiquitylation.Nat Cell Biol. 2010 Dec;12(12):1124-6. doi: 10.1038/ncb1210-1124. Nat Cell Biol. 2010. PMID: 21124306
-
Ubiquitylation: E3 ligases team up.Nat Rev Mol Cell Biol. 2011 Jan;12(1):6. doi: 10.1038/nrm3033. Epub 2010 Dec 8. Nat Rev Mol Cell Biol. 2011. PMID: 21139637 No abstract available.
References
-
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. - PubMed
-
- Turner GC, Du F, Varshavsky A. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature. 2000;405:579–583. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

