Slow conformational motions that favor sub-picosecond motions important for catalysis
- PMID: 21077591
- PMCID: PMC3018068
- DOI: 10.1021/jp1071296
Slow conformational motions that favor sub-picosecond motions important for catalysis
Abstract
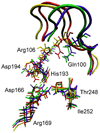
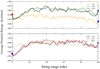
It has been accepted for many years that functionally important motions are crucial to binding properties of ligands in such molecules as hemoglobin and myoglobin. In enzymatic reactions, theory and now experiment are beginning to confirm the importance of motions on a fast (ps) time scale in the chemical step of the catalytic process. What is missing is a clear physical picture of how slow conformational fluctuations are related to the fast motions that have been identified as crucial. This paper presents a theoretical analysis of this issue for human heart lactate dehydrogenase. We will examine how slow conformational motions bring the system to conformations that are distinguished as catalytically competent because they favor specific fast motions.
Figures


References
-
- Warshel A, Sharma, Kato M, Y X, Liu H, Olsson M. Chem. Rev. 2006;106:3210–3235. - PubMed
-
- Henzler-Wildman K, Lei M, Thai V, Kerns S, Karplus M, Kern D. Nature. 2007;450:913–916. - PubMed
- Henzler-Wildman K, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson M, Petsko G, Karplus M, Hübner C, Kern D. Nature. 2007;450:838–844. - PubMed
- Agarwal P, Geist A, Gorin A. Biochemistry. 2004;43:10605–10618. - PubMed
- Agarwal P. J. Am. Chem. Soc. 2005;127:15248–15256. - PubMed
-
- Boehr D, Dyson H, Wright P. Chem. Rev. 2006;106:3055–3079. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

