GRAIL: a unique mediator of CD4 T-lymphocyte unresponsiveness
- PMID: 21078124
- PMCID: PMC3058357
- DOI: 10.1111/j.1742-4658.2010.07922.x
GRAIL: a unique mediator of CD4 T-lymphocyte unresponsiveness
Abstract
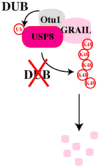
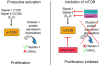
GRAIL (gene related to anergy in lymphocytes, also known as RNF128), an ubiquitin-protein ligase (E3), utilizes a unique single transmembrane protein with a split-function motif, and is an important gatekeeper of T-cell unresponsiveness. Although it may play a role in other CD4 T-cell functions including activation, survival and differentiation, GRAIL is most well characterized as a negative regulator of T-cell receptor responsiveness and cytokine production. Here, we review the recent literature on this remarkable E3 in the regulation of human and mouse CD4 T-cell unresponsiveness.
© 2010 The Authors Journal compilation © 2010 FEBS.
Figures
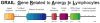
References
-
- Mueller DL. Mechanisms maintaining peripheral tolerance. Nat Immunol. 2010;11:21–27. - PubMed
-
- Schwartz RH. T cell anergy. Annu Rev Immunol. 2003;21:305–334. - PubMed
-
- Fathman CG, Lineberry NB. Molecular mechanisms of CD4+ T-cell anergy. Nat Rev Immunol. 2007;7:599–609. - PubMed
-
- Heissmeyer V, Macian F, Im SH, Varma R, Feske S, Venuprasad K, Gu H, Liu YC, Dustin ML, Rao A. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat Immunol. 2004;5:255–265. - PubMed
-
- Macian F, Garcia-Cozar F, Im SH, Horton HF, Byrne MC, Rao A. Transcriptional mechanisms underlying lymphocyte tolerance. Cell. 2002;109:719–731. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

